Crosstalk between Gut Microbiota and Host Immunity: Impact on Inflammation and Immunotherapy
Abstract
:1. Introduction
2. Role of Gut Microbiota and Their Metabolites in Neonatal Immune System Development
3. Interaction between Gut Microbiota and Host Innate Immune System
4. Interaction between Gut Microbiota and Adaptive Immune System
5. Crosstalk between Microbial Metabolites and Immune Regulation
5.1. Short-Chain Fatty Acids
5.2. Secondary Bile Acids
6. Influence of Environmental Microbiome Perturbation on the immune System
6.1. Antibiotic-Induced Microbiome Disturbances
6.2. Fecal Microbiota Transplantation
6.3. Diet-, Probiotic-, and Prebiotic-Induced Microbiome Alterations
6.3.1. High-Salt Diet
6.3.2. Dietary Polyphenols
6.3.3. Probiotics, Prebiotics, and Dietary Fiber
7. Dysregulation of Microbiome–Immunity Interaction in Various Diseases
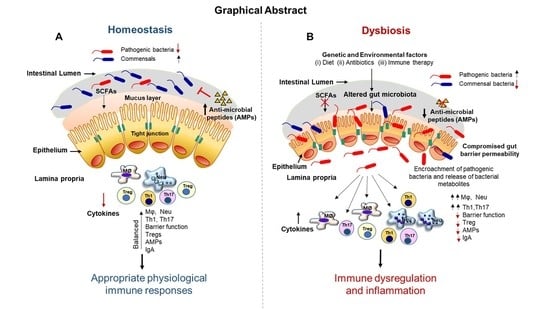
7.1. Gut Microbiota Dysbiosis and Immune Dysregulation
7.2. Gastrointestinal Infections
7.3. Inflammatory Bowel Diseases
7.4. Colorectal Carcinoma (CRC)
7.5. Hepatocellular Carcinoma (HCC)
7.6. Cardiovascular Disease
7.7. Diabetes
7.8. Hypertension
7.9. Rheumatoid Arthritis
7.10. Allergic Diseases
7.11. Psychiatric Disorders: The Gut–Brain Axis
8. Relationship between the Gut Microbiota and Their Metabolites in Immunotherapy
9. Promises, Challenges, and Risks in Immune–Microbiome Research
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GIT | Gastrointestinal tract |
| TLRs | Toll-like receptors |
| Treg | Foxp3+ regulatory T cells |
| Th17 | T helper 17 cells |
| PRRs | Pattern recognition receptors |
| PAMPs | Pathogen-associated molecular patterns |
| DCs | Dendritic cells |
| APCs | Antigen-presenting cells |
| SCFAs | Short-chain fatty acids |
| ILCs | Innate lymphoid cells |
| FMT | Fecal microbiota transplantation |
| IBD | Inflammatory bowel disease |
| NOD2 | Nucleotide-binding oligomerization domain-containing protein 2 |
| MDP | Muramyl dipeptide |
| TMAO | Trimethylamine-N-oxide |
| CRC | Colorectal carcinoma |
| HCC | Hepatocellular carcinoma |
| ICIs | Checkpoint inhibitors |
| CTLA-4 | Cytotoxic T lymphocyte antigen-4 |
| PD-1 | Programmed cell death protein 1 |
| PD-L1 | Programmed death ligand 1 |
| RA | Rheumatoid arthritis |
| LPS | Lipopolysaccharide |
| SFB | Segmented filamentous bacteria |
| CD | Crohn’s disease |
| UC | Ulcerative colitis |
| IEB | Intestinal–epithelial barrier |
| NAFLD | Non-alcoholic fatty liver disease |
| GPR | G-protein receptor |
| ROS | Reactive oxygen species |
| TNF | Tumor necrosis factor |
| NET | Neutrophil extracellular trap |
| NLR | NOD-like receptor |
| IEL | Intraepithelial lymphocytes |
| IL | Interleukin |
| RORγ | RAR-related orphan receptor gamma |
| Ig | Immunoglobulin |
| GPR | G-protein receptor |
| HDAC | Histone deacetylase |
| AhR | Aryl hydrocarbon receptor |
| HSD | High-salt diet |
| F/B ratio | Firmicutes/Bacteroidetes ratio |
| NaCl | Sodium chloride |
| CDI | Clostridium difficile infection |
| TGFβ | Tumor growth factor beta |
| CA | Cholic acid |
| DCA | Deoxycholic acid |
| UDCA | Ursodeoxycholic acid |
| LCA | Lithocholic acid |
| CVD | Cardiovascular diseases |
| TIDM | Type I diabetes mellitus |
| TIIDM | Type II diabetes mellitus |
References
- Donne, J. No Man Is an Island; Illustrated by Paul Peter Piech; Taurus Press: Willow Dene, UK, 1975. [Google Scholar]
- Beam, A.; Clinger, E.; Hao, L. Effect of Diet and Dietary Components on the Composition of the Gut Microbiota. Nutrients 2021, 13, 2795. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, F.; Ghosh, T.S.; O′Toole, P.W. The Healthy Microbiome-What Is the Definition of a Healthy Gut Microbiome? Gastroenterology 2021, 160, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. 2014, 13, 17–22. [Google Scholar]
- Sacks, D.; Baxter, B.; Campbell, B.C.; Carpenter, J.S.; Cognard, C.; Dippel, D.; Eesa, M.; Fischer, U.; Hausegger, K.; Hirsch, J.A. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. Int. J. Stroke 2018, 13, 612–632. [Google Scholar] [CrossRef] [Green Version]
- Matijasic, M.; Mestrovic, T.; Paljetak, H.C.; Peric, M.; Baresic, A.; Verbanac, D. Gut Microbiota beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int J. Mol. Sci. 2020, 21, 2668. [Google Scholar] [CrossRef] [Green Version]
- Giuffre, M.; Campigotto, M.; Campisciano, G.; Comar, M.; Croce, L.S. A story of liver and gut microbes: How does the intestinal flora affect liver disease? A review of the literature. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G889–G906. [Google Scholar] [CrossRef]
- Gomaa, E.Z. Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek 2020, 113, 2019–2040. [Google Scholar] [CrossRef]
- Mills, S.; Stanton, C.; Lane, J.A.; Smith, G.J.; Ross, R.P. Precision Nutrition and the Microbiome, Part I: Current State of the Science. Nutrients 2019, 11, 923. [Google Scholar] [CrossRef] [Green Version]
- Adak, A.; Khan, M.R. An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 2019, 76, 473–493. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Giuffre, M.; Moretti, R.; Campisciano, G.; da Silveira, A.B.M.; Monda, V.M.; Comar, M.; Di Bella, S.; Antonello, R.M.; Luzzati, R.; Croce, L.S. You Talking to Me? Says the Enteric Nervous System (ENS) to the Microbe. How Intestinal Microbes Interact with the ENS. J. Clin. Med. 2020, 9, 3705. [Google Scholar] [CrossRef]
- Cho, Y.; Shore, S.A. Obesity, Asthma, and the Microbiome. Physiology 2016, 31, 108–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McComb, S.; Thiriot, A.; Akache, B.; Krishnan, L.; Stark, F. Introduction to the Immune System. Methods Mol. Biol. 2019, 2024, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.; Karimi, N.; Morovati, S.; Alizadeh, K.; Kakish, J.E.; Vanderkamp, S.; Fazel, F.; Napoleoni, C.; Alizadeh, K.; Mehrani, Y.; et al. The Roles of Neutrophils in Cytokine Storms. Viruses 2021, 13, 2318. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, C.M.; Mazzoni, C.; Hogstrom, L.; Bryant, A.; Bergerat, A.; Cher, A.; Pochan, S.; Herman, P.; Carrigan, M.; Sharp, K.; et al. Delivery Mode Affects Stability of Early Infant Gut Microbiota. Cell Rep. Med. 2020, 1, 100156. [Google Scholar] [CrossRef]
- Selma-Royo, M.; Calatayud Arroyo, M.; Garcia-Mantrana, I.; Parra-Llorca, A.; Escuriet, R.; Martinez-Costa, C.; Collado, M.C. Perinatal environment shapes microbiota colonization and infant growth: Impact on host response and intestinal function. Microbiome 2020, 8, 167. [Google Scholar] [CrossRef]
- Subramanian, S.; Geng, H.; Du, C.; Chou, P.M.; Bu, H.F.; Wang, X.; Swaminathan, S.; Tan, S.C.; Ridlon, J.M.; De Plaen, I.G.; et al. Feeding mode influences dynamic gut microbiota signatures and affects susceptibility to anti-CD3 mAb-induced intestinal injury in neonatal mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2022, 323, G205–G218. [Google Scholar] [CrossRef]
- Wampach, L.; Heintz-Buschart, A.; Fritz, J.V.; Ramiro-Garcia, J.; Habier, J.; Herold, M.; Narayanasamy, S.; Kaysen, A.; Hogan, A.H.; Bindl, L.; et al. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat. Commun. 2018, 9, 5091. [Google Scholar] [CrossRef] [Green Version]
- Negi, S.; Hashimoto-Hill, S.; Alenghat, T. Neonatal microbiota-epithelial interactions that impact infection. Front. Microbiol. 2022, 13, 955051. [Google Scholar] [CrossRef] [PubMed]
- Senn, V.; Bassler, D.; Choudhury, R.; Scholkmann, F.; Righini-Grunder, F.; Vuille-Dit-Bile, R.N.; Restin, T. Microbial Colonization from the Fetus to Early Childhood-A Comprehensive Review. Front. Cell. Infect. Microbiol. 2020, 10, 573735. [Google Scholar] [CrossRef] [PubMed]
- Rackaityte, E.; Halkias, J.; Fukui, E.M.; Mendoza, V.F.; Hayzelden, C.; Crawford, E.D.; Fujimura, K.E.; Burt, T.D.; Lynch, S.V. Viable bacterial colonization is highly limited in the human intestine in utero. Nat. Med. 2020, 26, 599–607. [Google Scholar] [CrossRef]
- Li, Y.; Toothaker, J.M.; Ben-Simon, S.; Ozeri, L.; Schweitzer, R.; McCourt, B.T.; McCourt, C.C.; Werner, L.; Snapper, S.B.; Shouval, D.S.; et al. In utero human intestine harbors unique metabolome, including bacterial metabolites. JCI Insight 2020, 5, e138751. [Google Scholar] [CrossRef] [PubMed]
- Jain, N. The early life education of the immune system: Moms, microbes and (missed) opportunities. Gut Microbes 2020, 12, 1824564. [Google Scholar] [CrossRef]
- Gierynska, M.; Szulc-Dabrowska, L.; Struzik, J.; Mielcarska, M.B.; Gregorczyk-Zboroch, K.P. Integrity of the Intestinal Barrier: The Involvement of Epithelial Cells and Microbiota-A Mutual Relationship. Animals 2022, 12, 145. [Google Scholar] [CrossRef]
- Westrom, B.; Arevalo Sureda, E.; Pierzynowska, K.; Pierzynowski, S.G.; Perez-Cano, F.J. The Immature Gut Barrier and Its Importance in Establishing Immunity in Newborn Mammals. Front. Immunol. 2020, 11, 1153. [Google Scholar] [CrossRef]
- Rabe, H.; Lundell, A.C.; Sjoberg, F.; Ljung, A.; Strombeck, A.; Gio-Batta, M.; Maglio, C.; Nordstrom, I.; Andersson, K.; Nookaew, I.; et al. Neonatal gut colonization by Bifidobacterium is associated with higher childhood cytokine responses. Gut Microbes 2020, 12, 1847628. [Google Scholar] [CrossRef]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-mediated immune system imprinting early in life. Cell 2021, 184, 3884–3898. [Google Scholar] [CrossRef]
- Chin, N.; Mendez-Lagares, G.; Taft, D.H.; Laleau, V.; Kieu, H.; Narayan, N.R.; Roberts, S.B.; Mills, D.A.; Hartigan-O′Connor, D.J.; Flaherman, V.J. Transient Effect of Infant Formula Supplementation on the Intestinal Microbiota. Nutrients 2021, 13, 807. [Google Scholar] [CrossRef]
- Al Nabhani, Z.; Dulauroy, S.; Marques, R.; Cousu, C.; Al Bounny, S.; Dejardin, F.; Sparwasser, T.; Berard, M.; Cerf-Bensussan, N.; Eberl, G. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity 2019, 50, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Roubaud-Baudron, C.; Ruiz, V.E.; Swan, A.M., Jr.; Vallance, B.A.; Ozkul, C.; Pei, Z.; Li, J.; Battaglia, T.W.; Perez-Perez, G.I.; Blaser, M.J. Long-Term Effects of Early-Life Antibiotic Exposure on Resistance to Subsequent Bacterial Infection. mBio 2019, 10, e02820-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahenzli, J.; Koller, Y.; Wyss, M.; Geuking, M.B.; McCoy, K.D. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 2013, 14, 559–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Aidy, S.; Hooiveld, G.; Tremaroli, V.; Backhed, F.; Kleerebezem, M. The gut microbiota and mucosal homeostasis: Colonized at birth or at adulthood, does it matter? Gut Microbes 2013, 4, 118–124. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Li, Q.; Ren, J. Microbiota-Immune Interaction in the Pathogenesis of Gut-Derived Infection. Front. Immunol. 2019, 10, 1873. [Google Scholar] [CrossRef]
- McGuckin, M.A.; Linden, S.K.; Sutton, P.; Florin, T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011, 9, 265–278. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [Green Version]
- Minarrieta, L.; Ghorbani, P.; Sparwasser, T.; Berod, L. Metabolites: Deciphering the molecular language between DCs and their environment. Semin. Immunopathol. 2017, 39, 177–198. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Morikawa, M.; Tsujibe, S.; Kiyoshima-Shibata, J.; Watanabe, Y.; Kato-Nagaoka, N.; Shida, K.; Matsumoto, S. Microbiota of the Small Intestine Is Selectively Engulfed by Phagocytes of the Lamina Propria and Peyer′s Patches. PLoS ONE 2016, 11, e0163607. [Google Scholar] [CrossRef] [Green Version]
- Kohler, A.; Delbauve, S.; Smout, J.; Torres, D.; Flamand, V. Very early-life exposure to microbiota-induced TNF drives the maturation of neonatal pre-cDC1. Gut 2021, 70, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Hansson, G.C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 2016, 16, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Humayun, M.; Ayuso, J.M.; Park, K.Y.; Martorelli Di Genova, B.; Skala, M.C.; Kerr, S.C.; Knoll, L.J.; Beebe, D.J. Innate immune cell response to host-parasite interaction in a human intestinal tissue microphysiological system. Sci. Adv. 2022, 8, eabm8012. [Google Scholar] [CrossRef] [PubMed]
- Artis, D.; Spits, H. The biology of innate lymphoid cells. Nature 2015, 517, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Lankelma, J.M.; van Vught, L.A.; Belzer, C.; Schultz, M.J.; van der Poll, T.; de Vos, W.M.; Wiersinga, W.J. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: A pilot study. Intensive Care Med. 2017, 43, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Fournier, B.M.; Parkos, C.A. The role of neutrophils during intestinal inflammation. Mucosal Immunol. 2012, 5, 354–366. [Google Scholar] [CrossRef] [Green Version]
- Loh, J.T.; Lee, K.G.; Lee, A.P.; Teo, J.K.H.; Lim, H.L.; Kim, S.S.; Tan, A.H.; Lam, K.P. DOK3 maintains intestinal homeostasis by suppressing JAK2/STAT3 signaling and S100a8/9 production in neutrophils. Cell Death Dis. 2021, 12, 1054. [Google Scholar] [CrossRef]
- Seo, D.H.; Che, X.; Kim, S.; Kim, D.H.; Ma, H.W.; Kim, J.H.; Kim, T.I.; Kim, W.H.; Kim, S.W.; Cheon, J.H. Triggering Receptor Expressed on Myeloid Cells-1 Agonist Regulates Intestinal Inflammation via Cd177(+) Neutrophils. Front. Immunol. 2021, 12, 650864. [Google Scholar] [CrossRef]
- Vong, L.; Yeung, C.W.; Pinnell, L.J.; Sherman, P.M. Adherent-invasive Escherichia coli Exacerbates Antibiotic-associated Intestinal Dysbiosis and Neutrophil Extracellular Trap Activation. Inflamm. Bowel Dis. 2016, 22, 42–54. [Google Scholar] [CrossRef] [Green Version]
- Delano, M.J.; Ward, P.A. The immune system′s role in sepsis progression, resolution, and long-term outcome. Immunol. Rev. 2016, 274, 330–353. [Google Scholar] [CrossRef] [Green Version]
- Apostolov, A.K.; Hamani, M.; Hernandez-Vargas, H.; Igalouzene, R.; Guyennon, A.; Fesneau, O.; Marie, J.C.; Soudja, S.M. Common and Exclusive Features of Intestinal Intraepithelial gammadelta T Cells and Other gammadelta T Cell Subsets. Immunohorizons 2022, 6, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.S.; Severson, K.M.; Vaishnava, S.; Behrendt, C.L.; Yu, X.; Benjamin, J.L.; Ruhn, K.A.; Hou, B.; DeFranco, A.L.; Yarovinsky, F.; et al. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc. Natl. Acad. Sci. USA 2011, 108, 8743–8748. [Google Scholar] [CrossRef] [Green Version]
- Holtmeier, W.; Kabelitz, D. Gammadelta T cells link innate and adaptive immune responses. Chem. Immunol. Allergy 2005, 86, 151–183. [Google Scholar] [CrossRef]
- Tyler, C.J.; McCarthy, N.E.; Lindsay, J.O.; Stagg, A.J.; Moser, B.; Eberl, M. Antigen-Presenting Human gammadelta T Cells Promote Intestinal CD4(+) T Cell Expression of IL-22 and Mucosal Release of Calprotectin. J. Immunol. 2017, 198, 3417–3425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, Y.; Shi, F.; Zhang, X.; Zhang, Y.; Bi, K.; Chen, X.; Li, L.; Diao, H. Phospholipid metabolites of the gut microbiota promote hypoxia-induced intestinal injury via CD1d-dependent gammadelta T cells. Gut Microbes 2022, 14, 2096994. [Google Scholar] [CrossRef] [PubMed]
- Tomasello, E.; Bedoui, S. Intestinal innate immune cells in gut homeostasis and immunosurveillance. Immunol. Cell Biol. 2013, 91, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, D.; Le Bourhis, L.; Sauneuf, B.; Dechartres, A.; Rousseau, C.; Ouaaz, F.; Milder, M.; Louis, D.; Chiche, J.D.; Mira, J.P.; et al. Specific MAIT cell behaviour among innate-like T lymphocytes in critically ill patients with severe infections. Intensive Care Med. 2014, 40, 192–201. [Google Scholar] [CrossRef]
- Andreu-Ballester, J.C.; Tormo-Calandin, C.; Garcia-Ballesteros, C.; Perez-Griera, J.; Amigo, V.; Almela-Quilis, A.; Ruiz del Castillo, J.; Penarroja-Otero, C.; Ballester, F. Association of gammadelta T cells with disease severity and mortality in septic patients. Clin. Vaccine Immunol. 2013, 20, 738–746. [Google Scholar] [CrossRef]
- Noble, A.; Pring, E.T.; Durant, L.; Man, R.; Dilke, S.M.; Hoyles, L.; James, S.A.; Carding, S.R.; Jenkins, J.T.; Knight, S.C. Altered immunity to microbiota, B cell activation and depleted gammadelta/resident memory T cells in colorectal cancer. Cancer Immunol. Immunother. 2022, 71, 2619–2629. [Google Scholar] [CrossRef]
- Suzuki, H.; Jeong, K.I.; Itoh, K.; Doi, K. Regional variations in the distributions of small intestinal intraepithelial lymphocytes in germ-free and specific pathogen-free mice. Exp. Mol. Pathol. 2002, 72, 230–235. [Google Scholar] [CrossRef]
- Bedoui, S.; Heath, W.R.; Mueller, S.N. CD4(+) T-cell help amplifies innate signals for primary CD8(+) T-cell immunity. Immunol. Rev. 2016, 272, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Belz, G.T.; Shortman, K.; Bevan, M.J.; Heath, W.R. CD8alpha+ dendritic cells selectively present MHC class I-restricted noncytolytic viral and intracellular bacterial antigens in vivo. J. Immunol. 2005, 175, 196–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sztein, M.B.; Bafford, A.C.; Salerno-Goncalves, R. Salmonella enterica serovar Typhi exposure elicits ex vivo cell-type-specific epigenetic changes in human gut cells. Sci. Rep. 2020, 10, 13581. [Google Scholar] [CrossRef]
- Becattini, S.; Littmann, E.R.; Seok, R.; Amoretti, L.; Fontana, E.; Wright, R.; Gjonbalaj, M.; Leiner, I.M.; Plitas, G.; Hohl, T.M.; et al. Enhancing mucosal immunity by transient microbiota depletion. Nat. Commun. 2020, 11, 4475. [Google Scholar] [CrossRef]
- De Wit, J.; Souwer, Y.; Jorritsma, T.; Klaasse Bos, H.; ten Brinke, A.; Neefjes, J.; van Ham, S.M. Antigen-specific B cells reactivate an effective cytotoxic T cell response against phagocytosed Salmonella through cross-presentation. PLoS ONE 2010, 5, e13016. [Google Scholar] [CrossRef] [PubMed]
- Schnell, A.; Huang, L.; Singer, M.; Singaraju, A.; Barilla, R.M.; Regan, B.M.L.; Bollhagen, A.; Thakore, P.I.; Dionne, D.; Delorey, T.M.; et al. Stem-like intestinal Th17 cells give rise to pathogenic effector T cells during autoimmunity. Cell 2021, 184, 6281–6298.e23. [Google Scholar] [CrossRef]
- Omenetti, S.; Bussi, C.; Metidji, A.; Iseppon, A.; Lee, S.; Tolaini, M.; Li, Y.; Kelly, G.; Chakravarty, P.; Shoaie, S.; et al. The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells. Immunity 2019, 51, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef] [Green Version]
- Tan, T.G.; Sefik, E.; Geva-Zatorsky, N.; Kua, L.; Naskar, D.; Teng, F.; Pasman, L.; Ortiz-Lopez, A.; Jupp, R.; Wu, H.J.; et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E8141–E8150. [Google Scholar] [CrossRef] [Green Version]
- Sano, T.; Kageyama, T.; Fang, V.; Kedmi, R.; Martinez, C.S.; Talbot, J.; Chen, A.; Cabrera, I.; Gorshko, O.; Kurakake, R.; et al. Redundant cytokine requirement for intestinal microbiota-induced Th17 cell differentiation in draining lymph nodes. Cell Rep. 2021, 36, 109608. [Google Scholar] [CrossRef]
- Irons, E.E.; Cortes Gomez, E.; Andersen, V.L.; Lau, J.T.Y. Bacterial colonization and TH17 immunity are shaped by intestinal sialylation in neonatal mice. Glycobiology 2022, 32, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Ang, Q.Y.; Nayak, R.R.; Bustion, A.E.; Sandy, M.; Zhang, B.; Upadhyay, V.; Pollard, K.S.; Lynch, S.V.; Turnbaugh, P.J. Human gut bacterial metabolism drives Th17 activation and colitis. Cell Host Microbe 2022, 30, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Clay, S.L.; Lavoie, S.L.; Bae, S.; Lang, J.K.; Fonseca-Pereira, D.; Rosinski, K.G.; Ou, N.; Glickman, J.N.; Garrett, W.S. Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression. Gut Microbes 2021, 13, 1987780. [Google Scholar] [CrossRef] [PubMed]
- Malchow, S.; Leventhal, D.S.; Lee, V.; Nishi, S.; Socci, N.D.; Savage, P.A. Aire Enforces Immune Tolerance by Directing Autoreactive T Cells into the Regulatory T Cell Lineage. Immunity 2016, 44, 1102–1113. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Fujikado, N.; Kolodin, D.; Benoist, C.; Mathis, D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science 2015, 348, 589–594. [Google Scholar] [CrossRef] [Green Version]
- Ohnmacht, C.; Park, J.H.; Cording, S.; Wing, J.B.; Atarashi, K.; Obata, Y.; Gaboriau-Routhiau, V.; Marques, R.; Dulauroy, S.; Fedoseeva, M.; et al. Mucosal Immunology. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015, 349, 989–993. [Google Scholar] [CrossRef]
- Ramanan, D.; Sefik, E.; Galvan-Pena, S.; Wu, M.; Yang, L.; Yang, Z.; Kostic, A.; Golovkina, T.V.; Kasper, D.L.; Mathis, D.; et al. An Immunologic Mode of Multigenerational Transmission Governs a Gut Treg Setpoint. Cell 2020, 181, 1276–1290. [Google Scholar] [CrossRef]
- Sefik, E.; Geva-Zatorsky, N.; Oh, S.; Konnikova, L.; Zemmour, D.; McGuire, A.M.; Burzyn, D.; Ortiz-Lopez, A.; Lobera, M.; Yang, J.; et al. Mucosal Immunology. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 2015, 349, 993–997. [Google Scholar] [CrossRef] [Green Version]
- Lyu, M.; Suzuki, H.; Kang, L.; Gaspal, F.; Zhou, W.; Goc, J.; Zhou, L.; Zhou, J.; Zhang, W.; Bank, J.R.I.L.C.; et al. ILC3s select microbiota-specific regulatory T cells to establish tolerance in the gut. Nature 2022, 610, 744–751. [Google Scholar] [CrossRef]
- Chai, J.N.; Peng, Y.; Rengarajan, S.; Solomon, B.D.; Ai, T.L.; Shen, Z.; Perry, J.S.A.; Knoop, K.A.; Tanoue, T.; Narushima, S.; et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci. Immunol. 2017, 2, eaal5068. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yang, M.; Tang, L.; Wang, F.; Huang, S.; Liu, S.; Lei, Y.; Wang, S.; Xie, Z.; Wang, W.; et al. TLR4 regulates RORgammat(+) regulatory T-cell responses and susceptibility to colon inflammation through interaction with Akkermansia muciniphila. Microbiome 2022, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Du, H.X.; Yue, S.Y.; Niu, D.; Liu, C.; Zhang, L.G.; Chen, J.; Chen, Y.; Guan, Y.; Hua, X.L.; Li, C.; et al. Gut Microflora Modulates Th17/Treg Cell Differentiation in Experimental Autoimmune Prostatitis via the Short-Chain Fatty Acid Propionate. Front. Immunol. 2022, 13, 915218. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Yin, X.; Liu, Y.; Yan, X.; Zhang, S.; Wang, X.; Lin, Z.; Zhou, X.; Gao, J.; Wang, Z.; et al. Gut Dysbiosis Contributes to the Imbalance of Treg and Th17 Cells in Graves′ Disease Patients by Propionic Acid. J. Clin. Endocrinol. Metab. 2020, 105, 3526–3547. [Google Scholar] [CrossRef] [PubMed]
- Abokor, A.A.; McDaniel, G.H.; Golonka, R.M.; Campbell, C.; Brahmandam, S.; Yeoh, B.S.; Joe, B.; Vijay-Kumar, M.; Saha, P. Immunoglobulin A, an Active Liaison for Host-Microbiota Homeostasis. Microorganisms 2021, 9, 2117. [Google Scholar] [CrossRef] [PubMed]
- Pabst, O.; Cerovic, V.; Hornef, M. Secretory IgA in the Coordination of Establishment and Maintenance of the Microbiota. Trends Immunol. 2016, 37, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, D.B.; Suzuki, K.; Fagarasan, S. Fostering of advanced mutualism with gut microbiota by Immunoglobulin A. Immunol. Rev. 2016, 270, 20–31. [Google Scholar] [CrossRef]
- Vergani, S.; Muleta, K.G.; Da Silva, C.; Doyle, A.; Kristiansen, T.A.; Sodini, S.; Krausse, N.; Montano, G.; Kotarsky, K.; Nakawesi, J.; et al. A self-sustaining layer of early-life-origin B cells drives steady-state IgA responses in the adult gut. Immunity 2022, 55, 1829–1842. [Google Scholar] [CrossRef]
- New, J.S.; Dizon, B.L.P.; Fucile, C.F.; Rosenberg, A.F.; Kearney, J.F.; King, R.G. Neonatal Exposure to Commensal-Bacteria-Derived Antigens Directs Polysaccharide-Specific B-1 B Cell Repertoire Development. Immunity 2020, 53, 172–186. [Google Scholar] [CrossRef]
- Usami, K.; Niimi, K.; Matsuo, A.; Suyama, Y.; Sakai, Y.; Sato, S.; Fujihashi, K.; Kiyono, H.; Uchino, S.; Furukawa, M.; et al. The gut microbiota induces Peyer′s-patch-dependent secretion of maternal IgA into milk. Cell Rep. 2021, 36, 109655. [Google Scholar] [CrossRef]
- Mantis, N.J.; Rol, N.; Corthesy, B. Secretory IgA′s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef] [Green Version]
- De Sablet, T.; Chassard, C.; Bernalier-Donadille, A.; Vareille, M.; Gobert, A.P.; Martin, C. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2009, 77, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Rea, M.C.; Clayton, E.; O′Connor, P.M.; Shanahan, F.; Kiely, B.; Ross, R.P.; Hill, C. Antimicrobial activity of lacticin 3,147 against clinical Clostridium difficile strains. J. Med. Microbiol. 2007, 56, 940–946. [Google Scholar] [CrossRef] [Green Version]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut Microbiota and Immune System Interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sun, M.; Chen, F.; Cao, A.T.; Liu, H.; Zhao, Y.; Huang, X.; Xiao, Y.; Yao, S.; Zhao, Q.; et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017, 10, 946–956. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, T.; Miyauchi, E.; Kanaya, T.; Kato, T.; Nakanishi, Y.; Watanabe, T.; Kitami, T.; Taida, T.; Sasaki, T.; Negishi, H.; et al. Acetate differentially regulates IgA reactivity to commensal bacteria. Nature 2021, 595, 560–564. [Google Scholar] [CrossRef]
- Mandaliya, D.K.; Patel, S.; Seshadri, S. The Combinatorial Effect of Acetate and Propionate on High-Fat Diet Induced Diabetic Inflammation or Metaflammation and T Cell Polarization. Inflammation 2021, 44, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Eviston, D.; Hsu, P.; Marino, E.; Chidgey, A.; Santner-Nanan, B.; Wong, K.; Richards, J.L.; Yap, Y.A.; Collier, F.; et al. Decreased maternal serum acetate and impaired fetal thymic and regulatory T cell development in preeclampsia. Nat. Commun. 2019, 10, 3031. [Google Scholar] [CrossRef] [Green Version]
- Marino, E.; Richards, J.L.; McLeod, K.H.; Stanley, D.; Yap, Y.A.; Knight, J.; McKenzie, C.; Kranich, J.; Oliveira, A.C.; Rossello, F.J.; et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 2017, 18, 552–562. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Kang, S.G.; Jannasch, A.H.; Cooper, B.; Patterson, J.; Kim, C.H. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015, 8, 80–93. [Google Scholar] [CrossRef] [Green Version]
- Canani, R.B.; Costanzo, M.D.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef]
- Zheng, L.; Kelly, C.J.; Battista, K.D.; Schaefer, R.; Lanis, J.M.; Alexeev, E.E.; Wang, R.X.; Onyiah, J.C.; Kominsky, D.J.; Colgan, S.P. Microbial-Derived Butyrate Promotes Epithelial Barrier Function through IL-10 Receptor-Dependent Repression of Claudin-2. J. Immunol. 2017, 199, 2976–2984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432–445. [Google Scholar] [CrossRef] [Green Version]
- Luu, M.; Weigand, K.; Wedi, F.; Breidenbend, C.; Leister, H.; Pautz, S.; Adhikary, T.; Visekruna, A. Regulation of the effector function of CD8(+) T cells by gut microbiota-derived metabolite butyrate. Sci. Rep. 2018, 8, 14430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Yu, T.; Huang, X.; Bilotta, A.J.; Xu, L.; Lu, Y.; Sun, J.; Pan, F.; Zhou, J.; Zhang, W.; et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 2020, 11, 4457. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, F.; Tian, M.; Zhang, X.; Jin, X.; Jiang, Y.; Sun, X.; Wang, Y.; Peng, P.; Liu, J.; Xia, C.; et al. Butyrate enhances CPT1A activity to promote fatty acid oxidation and iTreg differentiation. Proc. Natl. Acad. Sci. USA 2021, 118, e2014681118. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [Green Version]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Sanchez, H.N.; Moroney, J.B.; Gan, H.; Shen, T.; Im, J.L.; Li, T.; Taylor, J.R.; Zan, H.; Casali, P. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat. Commun. 2020, 11, 60. [Google Scholar] [CrossRef] [Green Version]
- Rajput, M.; Momin, T.; Singh, A.; Banerjee, S.; Villasenor, A.; Sheldon, J.; Paudel, P.; Rajput, R. Determining the association between gut microbiota and its metabolites with higher intestinal Immunoglobulin A response. Vet. Anim. Sci. 2023, 19, 100279. [Google Scholar] [CrossRef] [PubMed]
- Dupraz, L.; Magniez, A.; Rolhion, N.; Richard, M.L.; Da Costa, G.; Touch, S.; Mayeur, C.; Planchais, J.; Agus, A.; Danne, C.; et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal gammadelta T cells. Cell Rep. 2021, 36, 109332. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez-Perez, O.; Cruz-Ramon, V.; Chinchilla-Lopez, P.; Mendez-Sanchez, N. The Role of the Gut Microbiota in Bile Acid Metabolism. Ann. Hepatol. 2017, 16 (Suppl. 1), S21–S26. [Google Scholar] [CrossRef]
- Heddes, M.; Altaha, B.; Niu, Y.; Reitmeier, S.; Kleigrewe, K.; Haller, D.; Kiessling, S. The intestinal clock drives the microbiome to maintain gastrointestinal homeostasis. Nat. Commun. 2022, 13, 6068. [Google Scholar] [CrossRef]
- Bogatyrev, S.R.; Rolando, J.C.; Ismagilov, R.F. Self-reinoculation with fecal flora changes microbiota density and composition leading to an altered bile-acid profile in the mouse small intestine. Microbiome 2020, 8, 19. [Google Scholar] [CrossRef] [Green Version]
- Van Best, N.; Rolle-Kampczyk, U.; Schaap, F.G.; Basic, M.; Olde Damink, S.W.M.; Bleich, A.; Savelkoul, P.H.M.; von Bergen, M.; Penders, J.; Hornef, M.W. Bile acids drive the newborn′s gut microbiota maturation. Nat. Commun. 2020, 11, 3692. [Google Scholar] [CrossRef]
- Chen, M.L.; Takeda, K.; Sundrud, M.S. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol. 2019, 12, 851–861. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Wang, C.; Huang, X.; Yi, S.; Pan, S.; Zhang, Y.; Yuan, G.; Cao, Q.; Ye, X.; Li, H. Gut microbiota-mediated secondary bile acids regulate dendritic cells to attenuate autoimmune uveitis through TGR5 signaling. Cell Rep. 2021, 36, 109726. [Google Scholar] [CrossRef]
- Biagioli, M.; Carino, A.; Cipriani, S.; Francisci, D.; Marchiano, S.; Scarpelli, P.; Sorcini, D.; Zampella, A.; Fiorucci, S. The Bile Acid Receptor GPBAR1 Regulates the M1/M2 Phenotype of Intestinal Macrophages and Activation of GPBAR1 Rescues Mice from Murine Colitis. J. Immunol. 2017, 199, 718–733. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorsky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature 2020, 577, 410–415. [Google Scholar] [CrossRef]
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.B.; Guo, C.J.; Violante, S.; et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020, 581, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Paik, D.; Yao, L.; Zhang, Y.; Bae, S.; D′Agostino, G.D.; Zhang, M.; Kim, E.; Franzosa, E.A.; Avila-Pacheco, J.; Bisanz, J.E.; et al. Human gut bacteria produce Tau(Eta)17-modulating bile acid metabolites. Nature 2022, 603, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Pols, T.W.H.; Puchner, T.; Korkmaz, H.I.; Vos, M.; Soeters, M.R.; de Vries, C.J.M. Lithocholic acid controls adaptive immune responses by inhibition of Th1 activation through the Vitamin D receptor. PLoS ONE 2017, 12, e0176715. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.D.; Mohammed, Z.; Roland, M.M.; Chatzistamou, I.; Jolly, A.; Schoettmer, L.M.; Arroyo, M.; Kakar, K.; Tian, Y.; Patterson, A.; et al. Defective humoral immunity disrupts bile acid homeostasis which promotes inflammatory disease of the small bowel. Nat. Commun. 2022, 13, 525. [Google Scholar] [CrossRef]
- Yoshida, S.; Ide, K.; Takeuchi, M.; Kawakami, K. Prenatal and early-life antibiotic use and risk of childhood asthma: A retrospective cohort study. Pediatr. Allergy Immunol. 2018, 29, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Ahmadizar, F.; Vijverberg, S.J.H.; Arets, H.G.M.; de Boer, A.; Lang, J.E.; Garssen, J.; Kraneveld, A.; Maitland-van der Zee, A.H. Early-life antibiotic exposure increases the risk of developing allergic symptoms later in life: A meta-analysis. Allergy 2018, 73, 971–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kronman, M.P.; Zaoutis, T.E.; Haynes, K.; Feng, R.; Coffin, S.E. Antibiotic exposure and IBD development among children: A population-based cohort study. Pediatrics 2012, 130, e794–e803. [Google Scholar] [CrossRef] [Green Version]
- Tapiainen, T.; Koivusaari, P.; Brinkac, L.; Lorenzi, H.A.; Salo, J.; Renko, M.; Pruikkonen, H.; Pokka, T.; Li, W.; Nelson, K.; et al. Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Sci. Rep. 2019, 9, 10635. [Google Scholar] [CrossRef] [Green Version]
- Nyangahu, D.D.; Lennard, K.S.; Brown, B.P.; Darby, M.G.; Wendoh, J.M.; Havyarimana, E.; Smith, P.; Butcher, J.; Stintzi, A.; Mulder, N.; et al. Disruption of maternal gut microbiota during gestation alters offspring microbiota and immunity. Microbiome 2018, 6, 124. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.B.; Konya, T.; Persaud, R.R.; Guttman, D.S.; Chari, R.S.; Field, C.J.; Sears, M.R.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: A prospective cohort study. BJOG 2016, 123, 983–993. [Google Scholar] [CrossRef]
- Tormo-Badia, N.; Hakansson, A.; Vasudevan, K.; Molin, G.; Ahrne, S.; Cilio, C.M. Antibiotic treatment of pregnant non-obese diabetic mice leads to altered gut microbiota and intestinal immunological changes in the offspring. Scand. J. Immunol. 2014, 80, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.H.; Liu, D.; Li, H.D.; Zhu, D.P.; He, Y.; Hou, T.; Yu, J.L. Prenatal and postnatal antibiotic exposure influences the gut microbiota of preterm infants in neonatal intensive care units. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, M.K.; Wang, B.; Ahmadi, S.; Burnham, C.A.; Tarr, P.I.; Warner, B.B.; Dantas, G. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol. 2016, 1, 16024. [Google Scholar] [CrossRef] [Green Version]
- Clark, R.H.; Bloom, B.T.; Spitzer, A.R.; Gerstmann, D.R. Reported medication use in the neonatal intensive care unit: Data from a large national data set. Pediatrics 2006, 117, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Du, W.; Xiao, S.; Zeng, B.; She, X.; Liu, D.; Du, H.; Li, L.; Li, F.; Ai, Q.; et al. Colonization of fecal microbiota from patients with neonatal necrotizing enterocolitis exacerbates intestinal injury in germfree mice subjected to necrotizing enterocolitis-induction protocol via alterations in butyrate and regulatory T cells. J. Transl. Med. 2021, 19, 510. [Google Scholar] [CrossRef]
- Niu, X.; Daniel, S.; Kumar, D.; Ding, E.Y.; Savani, R.C.; Koh, A.Y.; Mirpuri, J. Transient neonatal antibiotic exposure increases susceptibility to late-onset sepsis driven by microbiota-dependent suppression of type 3 innate lymphoid cells. Sci. Rep. 2020, 10, 12974. [Google Scholar] [CrossRef]
- Lange, K.; Buerger, M.; Stallmach, A.; Bruns, T. Effects of Antibiotics on Gut Microbiota. Dig. Dis. 2016, 34, 260–268. [Google Scholar] [CrossRef]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef]
- Wei, W.; Li, J.; Liu, F.; Wu, M.; Xiong, K.; He, Q.; Zhang, B.; Deng, Y.; Li, Y. Alteration of intestinal microecology by oral antibiotics promotes oral squamous cell carcinoma development. Mol. Immunol. 2022, 149, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4554–4561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyama, N.; Sudo, N.; Sogawa, H.; Kubo, C. Antibiotic use during infancy promotes a shift in the T(H)1/T(H)2 balance toward T(H)2-dominant immunity in mice. J. Allergy Clin. Immunol. 2001, 107, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.; Steinmeyer, S.; Bonfield, M.; Peterson, L.; Wang, T.; Gray, J.; Lewkowich, I.; Xu, Y.; Du, Y.; Guo, M.; et al. The balance between protective and pathogenic immune responses to pneumonia in the neonatal lung is enforced by gut microbiota. Sci. Transl. Med. 2022, 14, eabl3981. [Google Scholar] [CrossRef]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, A.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589–594. [Google Scholar] [CrossRef] [Green Version]
- Reijnders, D.; Goossens, G.H.; Hermes, G.D.; Neis, E.P.; van der Beek, C.M.; Most, J.; Holst, J.J.; Lenaerts, K.; Kootte, R.S.; Nieuwdorp, M.; et al. Effects of Gut Microbiota Manipulation by Antibiotics on Host Metabolism in Obese Humans: A Randomized Double-Blind Placebo-Controlled Trial. Cell Metab. 2016, 24, 63–74. [Google Scholar] [CrossRef] [Green Version]
- Perez-Reytor, D.; Puebla, C.; Karahanian, E.; Garcia, K. Use of Short-Chain Fatty Acids for the Recovery of the Intestinal Epithelial Barrier Affected by Bacterial Toxins. Front. Physiol. 2021, 12, 650313. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Xu, Z.Z.; Chang, M.W.; Marotz, C.A.; Saghatelian, A.; Knight, R.; Panda, S. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 2018, 9, 2872. [Google Scholar] [CrossRef] [Green Version]
- Vrieze, A.; Out, C.; Fuentes, S.; Jonker, L.; Reuling, I.; Kootte, R.S.; van Nood, E.; Holleman, F.; Knaapen, M.; Romijn, J.A.; et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol. 2014, 60, 824–831. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, J.; Li, J.V.; Zhou, N.Y.; Tang, H.; Wang, Y. Gut microbiota composition modifies fecal metabolic profiles in mice. J. Proteome Res. 2013, 12, 2987–2999. [Google Scholar] [CrossRef]
- Green, J.E.; Davis, J.A.; Berk, M.; Hair, C.; Loughman, A.; Castle, D.; Athan, E.; Nierenberg, A.A.; Cryan, J.F.; Jacka, F.; et al. Efficacy and safety of fecal microbiota transplantation for the treatment of diseases other than Clostridium difficile infection: A systematic review and meta-analysis. Gut Microbes 2020, 12, 1854640. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Chen, R.; Zhang, B.; Zhang, S.; Khan, B.A.; Zhu, D.; Wu, Z.; Xiao, C.; Chen, B.; Chen, F.; et al. Fecal microbiota transplantation treatment of autoimmune-mediated type 1 diabetes mellitus. Front. Immunol. 2022, 13, 930872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zou, G.; Li, B.; Du, X.; Sun, Z.; Sun, Y.; Jiang, X. Fecal Microbiota Transplantation (FMT) Alleviates Experimental Colitis in Mice by Gut Microbiota Regulation. J. Microbiol. Biotechnol. 2020, 30, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Costello, S.P.; Hughes, P.A.; Waters, O.; Bryant, R.V.; Vincent, A.D.; Blatchford, P.; Katsikeros, R.; Makanyanga, J.; Campaniello, M.A.; Mavrangelos, C.; et al. Effect of Fecal Microbiota Transplantation on 8-Week Remission in Patients with Ulcerative Colitis: A Randomized Clinical Trial. JAMA 2019, 321, 156–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paramsothy, S.; Kamm, M.A.; Kaakoush, N.O.; Walsh, A.J.; van den Bogaerde, J.; Samuel, D.; Leong, R.W.L.; Connor, S.; Ng, W.; Paramsothy, R.; et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: A randomised placebo-controlled trial. Lancet 2017, 389, 1218–1228. [Google Scholar] [CrossRef]
- Leong, K.S.W.; Jayasinghe, T.N.; Wilson, B.C.; Derraik, J.G.B.; Albert, B.B.; Chiavaroli, V.; Svirskis, D.M.; Beck, K.L.; Conlon, C.A.; Jiang, Y.; et al. Effects of Fecal Microbiome Transfer in Adolescents with Obesity: The Gut Bugs Randomized Controlled Trial. JAMA Netw. Open 2020, 3, e2030415. [Google Scholar] [CrossRef] [PubMed]
- Badran, M.; Mashaqi, S.; Gozal, D. The gut microbiome as a target for adjuvant therapy in obstructive sleep apnea. Expert Opin. Ther. Targets 2020, 24, 1263–1282. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, Q.; Wang, Y.; Sun, A.; Lin, Y.; Jin, Y.; Li, X. Fecal microbiota transplantation from chronic unpredictable mild stress mice donors affects anxiety-like and depression-like behavior in recipient mice via the gut microbiota-inflammation-brain axis. Stress 2019, 22, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Zeng, B.; Liu, M.; Chen, J.; Pan, J.; Han, Y.; Liu, Y.; Cheng, K.; Zhou, C.; Wang, H.; et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci. Adv. 2019, 5, eaau8317. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.W.; Kuo, C.H.; Kuo, F.C.; Wang, Y.K.; Hsu, W.H.; Yu, F.J.; Hu, H.M.; Hsu, P.I.; Wang, J.Y.; Wu, D.C. Fecal microbiota transplantation: Review and update. J. Formos Med. Assoc. 2019, 118 (Suppl. 1), S23–S31. [Google Scholar] [CrossRef]
- Tzianabos, A.O.; Onderdonk, A.B.; Rosner, B.; Cisneros, R.L.; Kasper, D.L. Structural features of polysaccharides that induce intra-abdominal abscesses. Science 1993, 262, 416–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; McLoughlin, R.M.; Cobb, B.A.; Charrel-Dennis, M.; Zaleski, K.J.; Golenbock, D.; Tzianabos, A.O.; Kasper, D.L. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J. Exp. Med. 2006, 203, 2853–2863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzianabos, A.; Wang, J.Y.; Kasper, D.L. Biological chemistry of immunomodulation by zwitterionic polysaccharides. Carbohydr. Res. 2003, 338, 2531–2538. [Google Scholar] [CrossRef] [PubMed]
- Tzianabos, A.O.; Finberg, R.W.; Wang, Y.; Chan, M.; Onderdonk, A.B.; Jennings, H.J.; Kasper, D.L. T cells activated by zwitterionic molecules prevent abscesses induced by pathogenic bacteria. J. Biol. Chem. 2000, 275, 6733–6740. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Mei, Q.; Lou, L.; Huang, Z.; Fu, Y.; Fan, J.; Wang, J.; Yin, N.; Zheng, Y.; Lu, Y.; et al. Ulcerative Colitis in Response to Fecal Microbiota Transplantation via Modulation of Gut Microbiota and Th17/Treg Cell Balance. Cells 2022, 11, 1851. [Google Scholar] [CrossRef]
- Lee, J.; d’Aigle, J.; Atadja, L.; Quaicoe, V.; Honarpisheh, P.; Ganesh, B.P.; Hassan, A.; Graf, J.; Petrosino, J.; Putluri, N.; et al. Gut Microbiota-Derived Short-Chain Fatty Acids Promote Poststroke Recovery in Aged Mice. Circ. Res. 2020, 127, 453–465. [Google Scholar] [CrossRef]
- Brunse, A.; Offersen, S.M.; Mosegaard, J.J.; Deng, L.; Damborg, P.; Nielsen, D.S.; Sangild, P.T.; Thymann, T.; Nguyen, D.N. Enteral broad-spectrum antibiotics antagonize the effect of fecal microbiota transplantation in preterm pigs. Gut Microbes 2021, 13, 1849997. [Google Scholar] [CrossRef]
- De Leon, L.M.; Watson, J.B.; Kelly, C.R. Transient flare of ulcerative colitis after fecal microbiota transplantation for recurrent Clostridium difficile infection. Clin. Gastroenterol. Hepatol. 2013, 11, 1036–1038. [Google Scholar] [CrossRef]
- Viennois, E.; Merlin, D.; Gewirtz, A.T.; Chassaing, B. Dietary Emulsifier-Induced Low-Grade Inflammation Promotes Colon Carcinogenesis. Cancer Res. 2017, 77, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Mantrana, I.; Selma-Royo, M.; Alcantara, C.; Collado, M.C. Shifts on Gut Microbiota Associated to Mediterranean Diet Adherence and Specific Dietary Intakes on General Adult Population. Front. Microbiol. 2018, 9, 890. [Google Scholar] [CrossRef] [Green Version]
- Kahleova, H.; Rembert, E.; Alwarith, J.; Yonas, W.N.; Tura, A.; Holubkov, R.; Agnello, M.; Chutkan, R.; Barnard, N.D. Effects of a Low-Fat Vegan Diet on Gut Microbiota in Overweight Individuals and Relationships with Body Weight, Body Composition, and Insulin Sensitivity. A Randomized Clinical Trial. Nutrients 2020, 12, 2917. [Google Scholar] [CrossRef] [PubMed]
- Ford, A.L.; Nagulesapillai, V.; Piano, A.; Auger, J.; Girard, S.A.; Christman, M.; Tompkins, T.A.; Dahl, W.J. Microbiota Stability and Gastrointestinal Tolerance in Response to a High-Protein Diet with and without a Prebiotic, Probiotic, and Synbiotic: A Randomized, Double-Blind, Placebo-Controlled Trial in Older Women. J. Acad. Nutr. Diet. 2020, 120, 500–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agocs, R.; Sugar, D.; Szabo, A.J. Is too much salt harmful? Yes. Pediatr. Nephrol. 2020, 35, 1777–1785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, P.M.; De Palma, G.; Serkis, V.; Lu, J.; Louis-Auguste, M.P.; McCarville, J.L.; Verdu, E.F.; Collins, S.M.; Bercik, P. High salt diet exacerbates colitis in mice by decreasing Lactobacillus levels and butyrate production. Microbiome 2018, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mahler, A.; Balogh, A.; Marko, L.; et al. Salt-responsive gut commensal modulates T(H)17 axis and disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef]
- Wang, C.; Huang, Z.; Yu, K.; Ding, R.; Ye, K.; Dai, C.; Xu, X.; Zhou, G.; Li, C. High-Salt Diet Has a Certain Impact on Protein Digestion and Gut Microbiota: A Sequencing and Proteome Combined Study. Front. Microbiol. 2017, 8, 1838. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Luo, H.; Wang, J.; Tang, W.; Lu, J.; Wu, S.; Xiong, Z.; Yang, G.; Chen, Z.; Lan, T.; et al. Enteric dysbiosis-linked gut barrier disruption triggers early renal injury induced by chronic high salt feeding in mice. Exp. Mol. Med. 2017, 49, e370. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Perumal, N.; Yadav, P.K.; Pandey, R.P.; Chang, C.M.; Raj, V.S. Amoxicillin impact on pathophysiology induced by short term high salt diet in mice. Sci. Rep. 2022, 12, 19351. [Google Scholar] [CrossRef]
- Matthias, J.; Heink, S.; Picard, F.; Zeitrag, J.; Kolz, A.; Chao, Y.Y.; Soll, D.; de Almeida, G.P.; Glasmacher, E.; Jacobsen, I.D.; et al. Salt generates antiinflammatory Th17 cells but amplifies pathogenicity in proinflammatory cytokine microenvironments. J. Clin. Investig. 2020, 130, 4587–4600. [Google Scholar] [CrossRef]
- Monteleone, I.; Marafini, I.; Dinallo, V.; Di Fusco, D.; Troncone, E.; Zorzi, F.; Laudisi, F.; Monteleone, G. Sodium chloride-enriched Diet Enhanced Inflammatory Cytokine Production and Exacerbated Experimental Colitis in Mice. J. Crohn’s Colitis 2017, 11, 237–245. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Lu, C.; Chen, J.; Cui, G.; Wang, L.; Yu, T.; Yang, Y.; Wu, W.; Ding, Y.; Li, L.; et al. High salt diet stimulates gut Th17 response and exacerbates TNBS-induced colitis in mice. Oncotarget 2017, 8, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Kleinewietfeld, M.; Manzel, A.; Titze, J.; Kvakan, H.; Yosef, N.; Linker, R.A.; Muller, D.N.; Hafler, D.A. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 2013, 496, 518–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manach, C.; Williamson, G.; Morand, C.; Scalbert, A.; Remesy, C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am. J. Clin. Nutr. 2005, 81, 230S–242S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzounis, X.; Vulevic, J.; Kuhnle, G.G.; George, T.; Leonczak, J.; Gibson, G.R.; Kwik-Uribe, C.; Spencer, J.P. Flavanol monomer-induced changes to the human faecal microflora. Br. J. Nutr. 2008, 99, 782–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; de Bruijn, W.J.C.; Bruins, M.E.; Vincken, J.P. Reciprocal Interactions between Epigallocatechin-3-gallate (EGCG) and Human Gut Microbiota In Vitro. J. Agric. Food Chem. 2020, 68, 9804–9815. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, S.; Li, T.; Li, N.; Han, D.; Zhang, B.; Xu, Z.Z.; Zhang, S.; Pang, J.; Wang, S.; et al. Gut microbiota from green tea polyphenol-dosed mice improves intestinal epithelial homeostasis and ameliorates experimental colitis. Microbiome 2021, 9, 184. [Google Scholar] [CrossRef]
- Sun, X.; Dey, P.; Bruno, R.S.; Zhu, J. EGCG and catechin relative to green tea extract differentially modulate the gut microbial metabolome and liver metabolome to prevent obesity in mice fed a high-fat diet. J. Nutr. Biochem. 2022, 109, 109094. [Google Scholar] [CrossRef]
- Dey, P.; Olmstead, B.D.; Sasaki, G.Y.; Vodovotz, Y.; Yu, Z.; Bruno, R.S. Epigallocatechin gallate but not catechin prevents nonalcoholic steatohepatitis in mice similar to green tea extract while differentially affecting the gut microbiota. J. Nutr. Biochem. 2020, 84, 108455. [Google Scholar] [CrossRef]
- Remely, M.; Ferk, F.; Sterneder, S.; Setayesh, T.; Roth, S.; Kepcija, T.; Noorizadeh, R.; Rebhan, I.; Greunz, M.; Beckmann, J.; et al. EGCG Prevents High Fat Diet-Induced Changes in Gut Microbiota, Decreases of DNA Strand Breaks, and Changes in Expression and DNA Methylation of Dnmt1 and MLH1 in C57BL/6J Male Mice. Oxid. Med. Cell. Longev. 2017, 2017, 3079148. [Google Scholar] [CrossRef] [Green Version]
- Cai, S.; Xie, L.W.; Xu, J.Y.; Zhou, H.; Yang, C.; Tang, L.F.; Tian, Y.; Li, M. (-)-Epigallocatechin-3-Gallate (EGCG) Modulates the Composition of the Gut Microbiota to Protect Against Radiation-Induced Intestinal Injury in Mice. Front. Oncol. 2022, 12, 848107. [Google Scholar] [CrossRef]
- Wu, Z.; Shen, J.; Xu, Q.; Xiang, Q.; Chen, Y.; Lv, L.; Zheng, B.; Wang, Q.; Wang, S.; Li, L. Epigallocatechin-3-Gallate Improves Intestinal Gut Microbiota Homeostasis and Ameliorates Clostridioides difficile Infection. Nutrients 2022, 14, 3756. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, J.; Wang, Y.; Li, T.; Zhao, J.; Zhang, C. Green tea polyphenols function as prooxidants to inhibit Pseudomonas aeruginosa and induce the expression of oxidative stress-related genes. Folia Microbiol. 2013, 58, 211–217. [Google Scholar] [CrossRef]
- Cui, Y.; Oh, Y.J.; Lim, J.; Youn, M.; Lee, I.; Pak, H.K.; Park, W.; Jo, W.; Park, S. AFM study of the differential inhibitory effects of the green tea polyphenol (-)-epigallocatechin-3-gallate (EGCG) against Gram-positive and Gram-negative bacteria. Food Microbiol. 2012, 29, 80–87. [Google Scholar] [CrossRef]
- Navarro-Martinez, M.D.; Navarro-Peran, E.; Cabezas-Herrera, J.; Ruiz-Gomez, J.; Garcia-Canovas, F.; Rodriguez-Lopez, J.N. Antifolate activity of epigallocatechin gallate against Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2005, 49, 2914–2920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.M.; Rock, C.O. Evaluation of epigallocatechin gallate and related plant polyphenols as inhibitors of the FabG and FabI reductases of bacterial type II fatty-acid synthase. J. Biol. Chem 2004, 279, 30994–31001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, L.G.; Chen, Y.J.; Tong, J.W.; Huang, J.A.; Li, J.; Gong, Y.S.; Liu, Z.H. Tea polyphenol epigallocatechin gallate inhibits Escherichia coli by increasing endogenous oxidative stress. Food Chem. 2017, 217, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Plamada, D.; Vodnar, D.C. Polyphenols-Gut Microbiota Interrelationship: A Transition to a New Generation of Prebiotics. Nutrients 2021, 14, 137. [Google Scholar] [CrossRef]
- Cristofori, F.; Dargenio, V.N.; Dargenio, C.; Miniello, V.L.; Barone, M.; Francavilla, R. Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation: A Door to the Body. Front. Immunol. 2021, 12, 578386. [Google Scholar] [CrossRef]
- Compare, D.; Rocco, A.; Coccoli, P.; Angrisani, D.; Sgamato, C.; Iovine, B.; Salvatore, U.; Nardone, G. Lactobacillus casei DG and its postbiotic reduce the inflammatory mucosal response: An ex-vivo organ culture model of post-infectious irritable bowel syndrome. BMC Gastroenterol. 2017, 17, 53. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, W.; Malinczak, C.A.; Fujimura, K.; Li, D.; McCauley, K.; Li, J.; Best, S.K.K.; Zhu, D.; Rasky, A.J.; Johnson, C.C.; et al. Maternal gut microbiome regulates immunity to RSV infection in offspring. J. Exp. Med. 2021, 218, e20210235. [Google Scholar] [CrossRef]
- Liu, H.Y.; Giraud, A.; Seignez, C.; Ahl, D.; Guo, F.; Sedin, J.; Walden, T.; Oh, J.H.; van Pijkeren, J.P.; Holm, L.; et al. Distinct B cell subsets in Peyer′s patches convey probiotic effects by Limosilactobacillus reuteri. Microbiome 2021, 9, 198. [Google Scholar] [CrossRef]
- So, D.; Whelan, K.; Rossi, M.; Morrison, M.; Holtmann, G.; Kelly, J.T.; Shanahan, E.R.; Staudacher, H.M.; Campbell, K.L. Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2018, 107, 965–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spinler, J.K.; Oezguen, N.; Runge, J.K.; Luna, R.A.; Karri, V.; Yang, J.; Hirschi, K.D. Dietary impact of a plant-derived microRNA on the gut microbiome. ExRNA 2020, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Wang, L.; Gu, Y.; Hou, H.; Liu, T.; Ding, Y.; Cao, H. Dietary Patterns and Gut Microbiota Changes in Inflammatory Bowel Disease: Current Insights and Future Challenges. Nutrients 2022, 14, 4003. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Quigley, E.M.M. Prebiotics and Probiotics in Digestive Health. Clin. Gastroenterol. Hepatol. 2019, 17, 333–344. [Google Scholar] [CrossRef]
- Cukrowska, B.; Ceregra, A.; Maciorkowska, E.; Surowska, B.; Zegadlo-Mylik, M.A.; Konopka, E.; Trojanowska, I.; Zakrzewska, M.; Bierla, J.B.; Zakrzewski, M.; et al. The Effectiveness of Probiotic Lactobacillus rhamnosus and Lactobacillus casei Strains in Children with Atopic Dermatitis and Cow′s Milk Protein Allergy: A Multicenter, Randomized, Double Blind, Placebo Controlled Study. Nutrients 2021, 13, 1169. [Google Scholar] [CrossRef] [PubMed]
- Cukrowska, B.; Bierla, J.B.; Zakrzewska, M.; Klukowski, M.; Maciorkowska, E. The Relationship between the Infant Gut Microbiota and Allergy. The Role of Bifidobacterium breve and Prebiotic Oligosaccharides in the Activation of Anti-Allergic Mechanisms in Early Life. Nutrients 2020, 12, 946. [Google Scholar] [CrossRef] [Green Version]
- Ho, J.; Nicolucci, A.C.; Virtanen, H.; Schick, A.; Meddings, J.; Reimer, R.A.; Huang, C. Effect of Prebiotic on Microbiota, Intestinal Permeability, and Glycemic Control in Children with Type 1 Diabetes. J. Clin. Endocrinol. Metab. 2019, 104, 4427–4440. [Google Scholar] [CrossRef] [Green Version]
- Kijmanawat, A.; Panburana, P.; Reutrakul, S.; Tangshewinsirikul, C. Effects of probiotic supplements on insulin resistance in gestational diabetes mellitus: A double-blind randomized controlled trial. J. Diabetes Investig. 2019, 10, 163–170. [Google Scholar] [CrossRef] [Green Version]
- Westaway, J.A.F.; Huerlimann, R.; Kandasamy, Y.; Miller, C.M.; Norton, R.; Watson, D.; Infante-Vilamil, S.; Rudd, D. Exploring the long-term colonisation and persistence of probiotic-prophylaxis species on the gut microbiome of preterm infants: A pilot study. Eur. J. Pediatr. 2022, 181, 3389–3400. [Google Scholar] [CrossRef] [PubMed]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, S.; Razazan, A.; Nagpal, R.; Jain, S.; Wang, B.; Mishra, S.P.; Wang, S.; Justice, J.; Ding, J.; McClain, D.A.; et al. Metformin Reduces Aging-Related Leaky Gut and Improves Cognitive Function by Beneficially Modulating Gut Microbiome/Goblet Cell/Mucin Axis. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, e9–e21. [Google Scholar] [CrossRef] [PubMed]
- Stevens, B.R.; Goel, R.; Seungbum, K.; Richards, E.M.; Holbert, R.C.; Pepine, C.J.; Raizada, M.K. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut 2018, 67, 1555–1557. [Google Scholar] [CrossRef]
- Thevaranjan, N.; Puchta, A.; Schulz, C.; Naidoo, A.; Szamosi, J.C.; Verschoor, C.P.; Loukov, D.; Schenck, L.P.; Jury, J.; Foley, K.P.; et al. Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host Microbe 2017, 21, 455–466. [Google Scholar] [CrossRef] [Green Version]
- Hamer, H.M.; De Preter, V.; Windey, K.; Verbeke, K. Functional analysis of colonic bacterial metabolism: Relevant to health? Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G1–G9. [Google Scholar] [CrossRef]
- McCall, I.C.; Betanzos, A.; Weber, D.A.; Nava, P.; Miller, G.W.; Parkos, C.A. Effects of phenol on barrier function of a human intestinal epithelial cell line correlate with altered tight junction protein localization. Toxicol. Appl. Pharmacol. 2009, 241, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, M.C.; Haak, B.W.; Hugenholtz, F.; Wiersinga, W.J. Gut microbiota and host defense in critical illness. Curr. Opin. Crit. Care 2017, 23, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Van der Poll, T.; van de Veerdonk, F.L.; Scicluna, B.P.; Netea, M.G. The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol. 2017, 17, 407–420. [Google Scholar] [CrossRef]
- Singer, J.R.; Blosser, E.G.; Zindl, C.L.; Silberger, D.J.; Conlan, S.; Laufer, V.A.; DiToro, D.; Deming, C.; Kumar, R.; Morrow, C.D.; et al. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat. Med. 2019, 25, 1772–1782. [Google Scholar] [CrossRef] [PubMed]
- Tovaglieri, A.; Sontheimer-Phelps, A.; Geirnaert, A.; Prantil-Baun, R.; Camacho, D.M.; Chou, D.B.; Jalili-Firoozinezhad, S.; de Wouters, T.; Kasendra, M.; Super, M.; et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 2019, 7, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.; Chang, E.B. Inflammatory Bowel Diseases (IBD) and the Microbiome-Searching the Crime Scene for Clues. Gastroenterology 2021, 160, 524–537. [Google Scholar] [CrossRef]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552. [Google Scholar] [CrossRef] [PubMed]
- Hale, V.L.; Chen, J.; Johnson, S.; Harrington, S.C.; Yab, T.C.; Smyrk, T.C.; Nelson, H.; Boardman, L.A.; Druliner, B.R.; Levin, T.R.; et al. Shifts in the Fecal Microbiota Associated with Adenomatous Polyps. Cancer Epidemiol. Biomark. Prev. 2017, 26, 85–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeoh, Y.K.; Chen, Z.; Wong, M.C.S.; Hui, M.; Yu, J.; Ng, S.C.; Sung, J.J.Y.; Chan, F.K.L.; Chan, P.K.S. Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut 2020, 69, 1998–2007. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.S.; Lee, F.Y.; Lee, S.D.; Tsai, Y.T.; Lin, H.C.; Lu, R.H.; Hsu, W.C.; Huang, C.C.; Wang, S.S.; Lo, K.J. Endotoxemia in patients with chronic liver diseases: Relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J. Hepatol 1995, 22, 165–172. [Google Scholar] [CrossRef]
- Singh, V.; Yeoh, B.S.; Chassaing, B.; Xiao, X.; Saha, P.; Aguilera Olvera, R.; Lapek, J.D., Jr.; Zhang, L.; Wang, W.B.; Hao, S.; et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 2018, 175, 679–694. [Google Scholar] [CrossRef] [Green Version]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef] [Green Version]
- Guasch-Ferre, M.; Hu, F.B.; Ruiz-Canela, M.; Bullo, M.; Toledo, E.; Wang, D.D.; Corella, D.; Gomez-Gracia, E.; Fiol, M.; Estruch, R.; et al. Plasma Metabolites from Choline Pathway and Risk of Cardiovascular Disease in the PREDIMED (Prevention with Mediterranean Diet) Study. J. Am. Heart Assoc. 2017, 6, e006524. [Google Scholar] [CrossRef] [PubMed]
- Millard, H.R.; Musani, S.K.; Dibaba, D.T.; Talegawkar, S.A.; Taylor, H.A.; Tucker, K.L.; Bidulescu, A. Dietary choline and betaine; associations with subclinical markers of cardiovascular disease risk and incidence of CVD, coronary heart disease and stroke: The Jackson Heart Study. Eur. J. Nutr. 2018, 57, 51–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013, 2, e01202. [Google Scholar] [CrossRef] [PubMed]
- Fazlollahi, M.; Chun, Y.; Grishin, A.; Wood, R.A.; Burks, A.W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.A.; Sicherer, S.H.; et al. Early-life gut microbiome and egg allergy. Allergy 2018, 73, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Bunyavanich, S.; Shen, N.; Grishin, A.; Wood, R.; Burks, W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.; Sicherer, S.; et al. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016, 138, 1122–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osbelt, L.; Wende, M.; Almasi, E.; Derksen, E.; Muthukumarasamy, U.; Lesker, T.R.; Galvez, E.J.C.; Pils, M.C.; Schalk, E.; Chhatwal, P.; et al. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe 2021, 29, 1663–1679. [Google Scholar] [CrossRef]
- Osbelt, L.; Thiemann, S.; Smit, N.; Lesker, T.R.; Schroter, M.; Galvez, E.J.C.; Schmidt-Hohagen, K.; Pils, M.C.; Muhlen, S.; Dersch, P.; et al. Variations in microbiota composition of laboratory mice influence Citrobacter rodentium infection via variable short-chain fatty acid production. PLoS Pathog. 2020, 16, e1008448. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, R.A.; Ng, K.M.; Correia, M.B.; Cabral, V.; Shi, H.; Sonnenburg, J.L.; Huang, K.C.; Xavier, K.B. Klebsiella michiganensis transmission enhances resistance to Enterobacteriaceae gut invasion by nutrition competition. Nat. Microbiol. 2020, 5, 630–641. [Google Scholar] [CrossRef] [Green Version]
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019, 83, e00007-19. [Google Scholar] [CrossRef]
- Jacobson, A.; Lam, L.; Rajendram, M.; Tamburini, F.; Honeycutt, J.; Pham, T.; Van Treuren, W.; Pruss, K.; Stabler, S.R.; Lugo, K.; et al. A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe 2018, 24, 296–307. [Google Scholar] [CrossRef] [Green Version]
- Izquierdo, M.; Lopez, J.; Gallardo, P.; Vidal, R.M.; Ossa, J.C.; Farfan, M.J. Bacteria from gut microbiota associated with diarrheal infections in children promote virulence of Shiga toxin-producing and enteroaggregative Escherichia coli pathotypes. Front. Cell. Infect. Microbiol. 2022, 12, 867205. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, N.A.; Schubert, A.M.; Flynn, K.J.; Leslie, J.L.; Sinani, H.; Bergin, I.L.; Young, V.B.; Schloss, P.D. The Gut Bacterial Community Potentiates Clostridioides difficile Infection Severity. mBio 2022, 13, e0118322. [Google Scholar] [CrossRef] [PubMed]
- Thiemann, S.; Smit, N.; Roy, U.; Lesker, T.R.; Galvez, E.J.C.; Helmecke, J.; Basic, M.; Bleich, A.; Goodman, A.L.; Kalinke, U.; et al. Enhancement of IFNgamma Production by Distinct Commensals Ameliorates Salmonella-Induced Disease. Cell Host Microbe 2017, 21, 682–694.e685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caballero, S.; Kim, S.; Carter, R.A.; Leiner, I.M.; Susac, B.; Miller, L.; Kim, G.J.; Ling, L.; Pamer, E.G. Cooperating Commensals Restore Colonization Resistance to Vancomycin-Resistant Enterococcus faecium. Cell Host Microbe 2017, 21, 592–602.e594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibbo, S.; Lopetuso, L.R.; Ianiro, G.; Di Rienzo, T.; Gasbarrini, A.; Cammarota, G. Role of microbiota and innate immunity in recurrent Clostridium difficile infection. J. Immunol. Res. 2014, 2014, 462740. [Google Scholar] [CrossRef] [Green Version]
- Warny, M.; Keates, A.C.; Keates, S.; Castagliuolo, I.; Zacks, J.K.; Aboudola, S.; Qamar, A.; Pothoulakis, C.; LaMont, J.T.; Kelly, C.P. p38 MAP kinase activation by Clostridium difficile toxin A mediates monocyte necrosis, IL-8 production, and enteritis. J. Clin. Investig. 2000, 105, 1147–1156. [Google Scholar] [CrossRef] [Green Version]
- Ishida, Y.; Maegawa, T.; Kondo, T.; Kimura, A.; Iwakura, Y.; Nakamura, S.; Mukaida, N. Essential involvement of IFN-gamma in Clostridium difficile toxin A-induced enteritis. J. Immunol. 2004, 172, 3018–3025. [Google Scholar] [CrossRef] [Green Version]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E., Jr.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 3114. [Google Scholar] [CrossRef] [Green Version]
- Rea, M.C.; Dobson, A.; O′Sullivan, O.; Crispie, F.; Fouhy, F.; Cotter, P.D.; Shanahan, F.; Kiely, B.; Hill, C.; Ross, R.P. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4639–4644. [Google Scholar] [CrossRef] [Green Version]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef] [Green Version]
- McDonald, L.C. Effects of short- and long-course antibiotics on the lower intestinal microbiome as they relate to traveller′s diarrhea. J. Travel Med. 2017, 24, S35–S38. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Deng, W.; Li, F.; Chen, Y.E.; Wang, P.U. Blockade of T helper 17 cell function ameliorates recurrent Clostridioides difficile infection in mice. Acta Biochim. Biophys. Sin. 2021, 53, 1290–1299. [Google Scholar] [CrossRef] [PubMed]
- Frisbee, A.L.; Saleh, M.M.; Young, M.K.; Leslie, J.L.; Simpson, M.E.; Abhyankar, M.M.; Cowardin, C.A.; Ma, J.Z.; Pramoonjago, P.; Turner, S.D.; et al. IL-33 drives group 2 innate lymphoid cell-mediated protection during Clostridium difficile infection. Nat. Commun. 2019, 10, 2712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Xu, Q.; Wang, Q.; Chen, Y.; Lv, L.; Zheng, B.; Yan, R.; Jiang, H.; Shen, J.; Wang, S.; et al. The impact of dietary fibers on Clostridioides difficile infection in a mouse model. Front. Cell. Infect. Microbiol. 2022, 12, 1028267. [Google Scholar] [CrossRef]
- Hayashi, A.; Nagao-Kitamoto, H.; Kitamoto, S.; Kim, C.H.; Kamada, N. The Butyrate-Producing Bacterium Clostridium butyricum Suppresses Clostridioides difficile Infection via Neutrophil- and Antimicrobial Cytokine-Dependent but GPR43/109a-Independent Mechanisms. J. Immunol. 2021, 206, 1576–1585. [Google Scholar] [CrossRef]
- Petrof, E.O.; Gloor, G.B.; Vanner, S.J.; Weese, S.J.; Carter, D.; Daigneault, M.C.; Brown, E.M.; Schroeter, K.; Allen-Vercoe, E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ′RePOOPulating′ the gut. Microbiome 2013, 1, 3. [Google Scholar] [CrossRef] [Green Version]
- Martz, S.L.; McDonald, J.A.; Sun, J.; Zhang, Y.G.; Gloor, G.B.; Noordhof, C.; He, S.M.; Gerbaba, T.K.; Blennerhassett, M.; Hurlbut, D.J.; et al. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci. Rep. 2015, 5, 16094. [Google Scholar] [CrossRef] [Green Version]
- Barbara, G.; Barbaro, M.R.; Fuschi, D.; Palombo, M.; Falangone, F.; Cremon, C.; Marasco, G.; Stanghellini, V. Inflammatory and Microbiota-Related Regulation of the Intestinal Epithelial Barrier. Front. Nutr. 2021, 8, 718356. [Google Scholar] [CrossRef]
- Chassaing, B.; Darfeuille-Michaud, A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 2011, 140, 1720–1728. [Google Scholar] [CrossRef]
- Swidsinski, A.; Ladhoff, A.; Pernthaler, A.; Swidsinski, S.; Loening-Baucke, V.; Ortner, M.; Weber, J.; Hoffmann, U.; Schreiber, S.; Dietel, M.; et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 2002, 122, 44–54. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.M.; Kemnitz, J.W. Age- and gender-related changes in body size, adiposity, and endocrine and metabolic parameters in free-ranging rhesus macaques. Am. J. Phys. Anthropol. 1992, 89, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vazquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn′s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, J.M.; de Zoete, M.R.; Palm, N.W.; Laenen, Y.; Bright, R.; Mallette, M.; Bu, K.; Bielecka, A.A.; Xu, F.; Hurtado-Lorenzo, A.; et al. Immunoglobulin A Targets a Unique Subset of the Microbiota in Inflammatory Bowel Disease. Cell Host Microbe 2021, 29, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Palm, N.W.; de Zoete, M.R.; Cullen, T.W.; Barry, N.A.; Stefanowski, J.; Hao, L.; Degnan, P.H.; Hu, J.; Peter, I.; Zhang, W.; et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014, 158, 1000–1010. [Google Scholar] [CrossRef] [Green Version]
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; von Bergen, M.; et al. Dysbiotic gut microbiota causes transmissible Crohn′s disease-like ileitis independent of failure in antimicrobial defence. Gut 2016, 65, 225–237. [Google Scholar] [CrossRef] [Green Version]
- Torres, J.; Hu, J.; Seki, A.; Eisele, C.; Nair, N.; Huang, R.; Tarassishin, L.; Jharap, B.; Cote-Daigneault, J.; Mao, Q.; et al. Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ-free mice. Gut 2020, 69, 42–51. [Google Scholar] [CrossRef] [Green Version]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef]
- Facchin, S.; Vitulo, N.; Calgaro, M.; Buda, A.; Romualdi, C.; Pohl, D.; Perini, B.; Lorenzon, G.; Marinelli, C.; D′Inca, R.; et al. Microbiota changes induced by microencapsulated sodium butyrate in patients with inflammatory bowel disease. Neurogastroenterol. Motil. 2020, 32, e13914. [Google Scholar] [CrossRef]
- Li, G.; Lin, J.; Zhang, C.; Gao, H.; Lu, H.; Gao, X.; Zhu, R.; Li, Z.; Li, M.; Liu, Z. Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes 2021, 13, 1968257. [Google Scholar] [CrossRef] [PubMed]
- Hartog, A.; Belle, F.N.; Bastiaans, J.; de Graaff, P.; Garssen, J.; Harthoorn, L.F.; Vos, A.P. A potential role for regulatory T-cells in the amelioration of DSS induced colitis by dietary non-digestible polysaccharides. J. Nutr. Biochem. 2015, 26, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Yeoh, B.S.; Walker, R.E.; Xiao, X.; Saha, P.; Golonka, R.M.; Cai, J.; Bretin, A.C.A.; Cheng, X.; Liu, Q.; et al. Microbiota fermentation-NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut 2019, 68, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xu, Y.; Wang, X.; Rao, L.; Yan, X.; Gao, R.; Shen, T.; Zhou, Y.; Kong, C.; Zhou, L. Probiotic Cocktail Alleviates Intestinal Inflammation Through Improving Gut Microbiota and Metabolites in Colitis Mice. Front. Cell. Infect. Microbiol. 2022, 12, 886061. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Shen, Y.; Cen, M.; Zhu, Y.; Cheng, F.; Tang, L.; Zheng, X.; Kim, J.J.; Dai, N.; Hu, W. Modulation of the Gut Microbiota-farnesoid X Receptor Axis Improves Deoxycholic Acid-induced Intestinal Inflammation in Mice. J. Crohn’s Colitis 2021, 15, 1197–1210. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Cen, M.; Shen, Y.; Zhu, Y.; Cheng, F.; Tang, L.; Hu, W.; Dai, N. Deoxycholic Acid-Induced Gut Dysbiosis Disrupts Bile Acid Enterohepatic Circulation and Promotes Intestinal Inflammation. Dig. Dis. Sci. 2021, 66, 568–576. [Google Scholar] [CrossRef]
- Liu, T.C.; Kern, J.T.; Jain, U.; Sonnek, N.M.; Xiong, S.; Simpson, K.F.; VanDussen, K.L.; Winkler, E.S.; Haritunians, T.; Malique, A.; et al. Western diet induces Paneth cell defects through microbiome alterations and farnesoid X receptor and type I interferon activation. Cell Host Microbe 2021, 29, 988–1001. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, J.; Ren, X.; Zhang, Y.; Ke, Z.; Zhou, J.; Wang, Y.; Zhang, Y.; Liu, Y. Cholecystectomy-induced secondary bile acids accumulation ameliorates colitis through inhibiting monocyte/macrophage recruitment. Gut Microbes 2022, 14, 2107387. [Google Scholar] [CrossRef]
- Van den Bossche, L.; Hindryckx, P.; Devisscher, L.; Devriese, S.; Van Welden, S.; Holvoet, T.; Vilchez-Vargas, R.; Vital, M.; Pieper, D.H.; Vanden Bussche, J.; et al. Ursodeoxycholic Acid and Its Taurine- or Glycine-Conjugated Species Reduce Colitogenic Dysbiosis and Equally Suppress Experimental Colitis in Mice. Appl. Environ. Microbiol. 2017, 83, e02766-16. [Google Scholar] [CrossRef] [Green Version]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Moltzau Anderson, J.; Lipinski, S.; Sommer, F.; Pan, W.H.; Boulard, O.; Rehman, A.; Falk-Paulsen, M.; Stengel, S.T.; Aden, K.; Hasler, R.; et al. NOD2 Influences Trajectories of Intestinal Microbiota Recovery After Antibiotic Perturbation. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 365–389. [Google Scholar] [CrossRef] [PubMed]
- Danne, C.; Michaudel, C.; Skerniskyte, J.; Planchais, J.; Magniez, A.; Agus, A.; Michel, M.L.; Lamas, B.; Da Costa, G.; Spatz, M.; et al. CARD9 in neutrophils protects from colitis and controls mitochondrial metabolism and cell survival. Gut 2022. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, S.; Conway, K.L.; Lassen, K.G.; Jijon, H.B.; Pan, H.; Chun, E.; Michaud, M.; Lang, J.K.; Gallini Comeau, C.A.; Dreyfuss, J.M.; et al. The Crohn′s disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response. Elife 2019, 8, e39982. [Google Scholar] [CrossRef] [PubMed]
- Aden, K.; Tran, F.; Ito, G.; Sheibani-Tezerji, R.; Lipinski, S.; Kuiper, J.W.; Tschurtschenthaler, M.; Saveljeva, S.; Bhattacharyya, J.; Hasler, R.; et al. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J. Exp. Med. 2018, 215, 2868–2886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.B.; Zhou, Y.L.; Fang, J.Y. Gut Microbiota in Cancer Immune Response and Immunotherapy. Trends Cancer 2021, 7, 647–660. [Google Scholar] [CrossRef]
- Xu, C.; Fan, L.; Lin, Y.; Shen, W.; Qi, Y.; Zhang, Y.; Chen, Z.; Wang, L.; Long, Y.; Hou, T.; et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes 2021, 13, 1980347. [Google Scholar] [CrossRef]
- Hussan, H.; Clinton, S.K.; Roberts, K.; Bailey, M.T. Fusobacterium′s link to colorectal neoplasia sequenced: A systematic review and future insights. World J. Gastroenterol. 2017, 23, 8626–8650. [Google Scholar] [CrossRef]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866. [Google Scholar] [CrossRef] [Green Version]
- Gur, C.; Maalouf, N.; Shhadeh, A.; Berhani, O.; Singer, B.B.; Bachrach, G.; Mandelboim, O. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology 2019, 8, e1581531. [Google Scholar] [CrossRef] [Green Version]
- Loesche, W.J.; Gibbons, R.J. Amino acid fermentation by Fusobacterium nucleatum. Arch. Oral Biol. 1968, 13, 191–202. [Google Scholar] [CrossRef]
- Lavoie, S.; Chun, E.; Bae, S.; Brennan, C.A.; Gallini Comeau, C.A.; Lang, J.K.; Michaud, M.; Hoveyda, H.R.; Fraser, G.L.; Fuller, M.H.; et al. Expression of Free Fatty Acid Receptor 2 by Dendritic Cells Prevents Their Expression of Interleukin 27 and Is Required for Maintenance of Mucosal Barrier and Immune Response Against Colorectal Tumors in Mice. Gastroenterology 2020, 158, 1359–1372. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.P.; et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014, 4, 1387–1397. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Liu, L.; Dou, Y.; Xu, M.; Liu, T.; Wang, S.; Zhang, Y.; Deng, B.; Wang, B.; Cao, H. Deoxycholic acid activates epidermal growth factor receptor and promotes intestinal carcinogenesis by ADAM17-dependent ligand release. J. Cell. Mol. Med. 2018, 22, 4263–4273. [Google Scholar] [CrossRef] [PubMed]
- Pai, R.; Tarnawski, A.S.; Tran, T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol. Biol. Cell 2004, 15, 2156–2163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, T.; Coulter, S.; Yoshihara, E.; Oh, T.G.; Fang, S.; Cayabyab, F.; Zhu, Q.; Zhang, T.; Leblanc, M.; Liu, S.; et al. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 2019, 176, 1098–1112. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Yang, M.; Dong, W.; Liu, T.; Song, X.; Gu, Y.; Wang, S.; Liu, Y.; Abla, Z.; Qiao, X.; et al. Gut Dysbiosis and Abnormal Bile Acid Metabolism in Colitis-Associated Cancer. Gastroenterol. Res. Pract. 2021, 2021, 6645970. [Google Scholar] [CrossRef]
- Ternes, D.; Tsenkova, M.; Pozdeev, V.I.; Meyers, M.; Koncina, E.; Atatri, S.; Schmitz, M.; Karta, J.; Schmoetten, M.; Heinken, A.; et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat. Metab. 2022, 4, 458–475. [Google Scholar] [CrossRef]
- Fan, L.; Xu, C.; Ge, Q.; Lin, Y.; Wong, C.C.; Qi, Y.; Ye, B.; Lian, Q.; Zhuo, W.; Si, J.; et al. A. muciniphila Suppresses Colorectal Tumorigenesis by Inducing TLR2/NLRP3-Mediated M1-Like TAMs. Cancer Immunol. Res. 2021, 9, 1111–1124. [Google Scholar] [CrossRef]
- Zegarra Ruiz, D.F.; Kim, D.V.; Norwood, K.; Saldana-Morales, F.B.; Kim, M.; Ng, C.; Callaghan, R.; Uddin, M.; Chang, L.C.; Longman, R.S.; et al. Microbiota manipulation to increase macrophage IL-10 improves colitis and limits colitis-associated colorectal cancer. Gut Microbes 2022, 14, 2119054. [Google Scholar] [CrossRef]
- Yu, A.I.; Zhao, L.; Eaton, K.A.; Ho, S.; Chen, J.; Poe, S.; Becker, J.; Gonzalez, A.; McKinstry, D.; Hasso, M.; et al. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Rep. 2020, 31, 107471. [Google Scholar] [CrossRef]
- Peuker, K.; Strigli, A.; Tauriello, D.V.F.; Hendricks, A.; von Schonfels, W.; Burmeister, G.; Brosch, M.; Herrmann, A.; Kruger, S.; Nitsche, J.; et al. Microbiota-dependent activation of the myeloid calcineurin-NFAT pathway inhibits B7H3- and B7H4-dependent anti-tumor immunity in colorectal cancer. Immunity 2022, 55, 701–717. [Google Scholar] [CrossRef] [PubMed]
- Overacre-Delgoffe, A.E.; Bumgarner, H.J.; Cillo, A.R.; Burr, A.H.P.; Tometich, J.T.; Bhattacharjee, A.; Bruno, T.C.; Vignali, D.A.A.; Hand, T.W. Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity 2021, 54, 2812–2824.e2814. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef]
- Park, E.J.; Lee, J.H.; Yu, G.Y.; He, G.; Ali, S.R.; Holzer, R.G.; Osterreicher, C.H.; Takahashi, H.; Karin, M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010, 140, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Said, I.; Ahad, H.; Said, A. Gut microbiome in non-alcoholic fatty liver disease associated hepatocellular carcinoma: Current knowledge and potential for therapeutics. World J. Gastrointest. Oncol. 2022, 14, 947–958. [Google Scholar] [CrossRef]
- Behary, J.; Raposo, A.E.; Amorim, N.M.L.; Zheng, H.; Gong, L.; McGovern, E.; Chen, J.; Liu, K.; Beretov, J.; Theocharous, C.; et al. Defining the temporal evolution of gut dysbiosis and inflammatory responses leading to hepatocellular carcinoma in Mdr2 −/− mouse model. BMC Microbiol. 2021, 21, 113. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Paroni Sterbini, F.; Petito, V.; et al. Hepatocellular Carcinoma Is Associated with Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef] [Green Version]
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020, 9, 4232–4250. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Li, F.; Zhuang, Y.; Xu, J.; Wang, J.; Mao, X.; Zhang, Y.; Liu, X. Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog. 2019, 11, 1. [Google Scholar] [CrossRef]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Grat, M.; Wronka, K.M.; Krasnodebski, M.; Masior, L.; Lewandowski, Z.; Kosinska, I.; Grat, K.; Stypulkowski, J.; Rejowski, S.; Wasilewicz, M.; et al. Profile of Gut Microbiota Associated with the Presence of Hepatocellular Cancer in Patients with Liver Cirrhosis. Transplant. Proc. 2016, 48, 1687–1691. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Hara, E. Gut-liver axis-mediated mechanism of liver cancer: A special focus on the role of gut microbiota. Cancer Sci. 2021, 112, 4433–4443. [Google Scholar] [CrossRef] [PubMed]
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut Microbiota Promotes Obesity-Associated Liver Cancer through PGE(2)-Mediated Suppression of Antitumor Immunity. Cancer Discov. 2017, 7, 522–538. [Google Scholar] [CrossRef] [Green Version]
- Golonka, R.M.; Xiao, X.; Abokor, A.A.; Joe, B.; Vijay-Kumar, M. Altered nutrient status reprograms host inflammation and metabolic health via gut microbiota. J. Nutr. Biochem. 2020, 80, 108360. [Google Scholar] [CrossRef] [PubMed]
- Hosseinkhani, F.; Heinken, A.; Thiele, I.; Lindenburg, P.W.; Harms, A.C.; Hankemeier, T. The contribution of gut bacterial metabolites in the human immune signaling pathway of non-communicable diseases. Gut Microbes 2021, 13, 1882927. [Google Scholar] [CrossRef]
- Yue, C.; Yang, X.; Li, J.; Chen, X.; Zhao, X.; Chen, Y.; Wen, Y. Trimethylamine N-oxide prime NLRP3 inflammasome via inhibiting ATG16L1-induced autophagy in colonic epithelial cells. Biochem. Biophys. Res. Commun. 2017, 490, 541–551. [Google Scholar] [CrossRef]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Guasti, L.; Galliazzo, S.; Molaro, M.; Visconti, E.; Pennella, B.; Gaudio, G.V.; Lupi, A.; Grandi, A.M.; Squizzato, A. TMAO as a biomarker of cardiovascular events: A systematic review and meta-analysis. Intern. Emerg. Med. 2021, 16, 201–207. [Google Scholar] [CrossRef]
- Fang, C.; Zuo, K.; Jiao, K.; Zhu, X.; Fu, Y.; Zhong, J.; Xu, L.; Yang, X. PAGln, an Atrial Fibrillation-Linked Gut Microbial Metabolite, Acts as a Promoter of Atrial Myocyte Injury. Biomolecules 2022, 12, 1120. [Google Scholar] [CrossRef]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.J.; Saad, S.; Tillett, B.J.; McGuire, H.M.; Bordbar, S.; Yap, Y.A.; Nguyen, L.T.; Wilkins, M.R.; Corley, S.; Brodie, S.; et al. Metabolite-based dietary supplementation in human type 1 diabetes is associated with microbiota and immune modulation. Microbiome 2022, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Girdhar, K.; Huang, Q.; Chow, I.T.; Vatanen, T.; Brady, C.; Raisingani, A.; Autissier, P.; Atkinson, M.A.; Kwok, W.W.; Kahn, C.R.; et al. A gut microbial peptide and molecular mimicry in the pathogenesis of type 1 diabetes. Proc. Natl. Acad. Sci. USA 2022, 119, e2120028119. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, S.; Sen, P.; Dickens, A.M.; Alves, M.A.; Harkonen, T.; Honkanen, J.; Vatanen, T.; Xavier, R.J.; Hyotylainen, T.; Knip, M.; et al. Dysregulation of secondary bile acid metabolism precedes islet autoimmunity and type 1 diabetes. Cell Rep. Med. 2022, 3, 100762. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Zhou, M.; Shao, J.; Wu, C.Y.; Shu, L.; Dong, W.; Liu, Y.; Chen, M.; Wynn, R.M.; Wang, J.; Wang, J.; et al. Targeting BCAA Catabolism to Treat Obesity-Associated Insulin Resistance. Diabetes 2019, 68, 1730–1746. [Google Scholar] [CrossRef] [Green Version]
- Bodogai, M.; O′Connell, J.; Kim, K.; Kim, Y.; Moritoh, K.; Chen, C.; Gusev, F.; Vaughan, K.; Shulzhenko, N.; Mattison, J.A.; et al. Commensal bacteria contribute to insulin resistance in aging by activating innate B1a cells. Sci. Transl. Med. 2018, 10, eaat4271. [Google Scholar] [CrossRef]
- Huang, J.P.; Cheng, M.L.; Hung, C.Y.; Wang, C.H.; Hsieh, P.S.; Shiao, M.S.; Chen, J.K.; Li, D.E.; Hung, L.M. Docosapentaenoic acid and docosahexaenoic acid are positively associated with insulin sensitivity in rats fed high-fat and high-fructose diets. J. Diabetes 2017, 9, 936–946. [Google Scholar] [CrossRef]
- Wang, H.; Lu, Y.; Yan, Y.; Tian, S.; Zheng, D.; Leng, D.; Wang, C.; Jiao, J.; Wang, Z.; Bai, Y. Promising Treatment for Type 2 Diabetes: Fecal Microbiota Transplantation Reverses Insulin Resistance and Impaired Islets. Front. Cell. Infect. Microbiol. 2019, 9, 455. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Zhang, C.; Wang, P.; Wei, W.; Shi, X.; Wang, P.; Yang, J.; Wang, L.; Tang, S.; Fang, Y.; et al. Cardiovascular Benefits of Empagliflozin Are Associated with Gut Microbiota and Plasma Metabolites in Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2022, 107, 1888–1896. [Google Scholar] [CrossRef]
- Montandon, S.A.; Jornayvaz, F.R. Effects of Antidiabetic Drugs on Gut Microbiota Composition. Genes 2017, 8, 250. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Manneras-Holm, L.; Stahlman, M.; Olsson, L.M.; Serino, M.; Planas-Felix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adnan, S.; Nelson, J.W.; Ajami, N.J.; Venna, V.R.; Petrosino, J.F.; Bryan, R.M., Jr.; Durgan, D.J. Alterations in the gut microbiota can elicit hypertension in rats. Physiol. Genom. 2017, 49, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Marques, F.Z.; Nelson, E.; Chu, P.Y.; Horlock, D.; Fiedler, A.; Ziemann, M.; Tan, J.K.; Kuruppu, S.; Rajapakse, N.W.; El-Osta, A.; et al. High-Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice. Circulation 2017, 135, 964–977. [Google Scholar] [CrossRef]
- Santisteban, M.M.; Qi, Y.; Zubcevic, J.; Kim, S.; Yang, T.; Shenoy, V.; Cole-Jeffrey, C.T.; Lobaton, G.O.; Stewart, D.C.; Rubiano, A.; et al. Hypertension-Linked Pathophysiological Alterations in the Gut. Circ. Res. 2017, 120, 312–323. [Google Scholar] [CrossRef]
- Karbach, S.H.; Schonfelder, T.; Brandao, I.; Wilms, E.; Hormann, N.; Jackel, S.; Schuler, R.; Finger, S.; Knorr, M.; Lagrange, J.; et al. Gut Microbiota Promote Angiotensin II-Induced Arterial Hypertension and Vascular Dysfunction. J. Am. Heart Assoc. 2016, 5, e003698. [Google Scholar] [CrossRef] [Green Version]
- Avery, E.G.; Bartolomaeus, H.; Rauch, A.; Chen, C.Y.; N′Diaye, G.; Lober, U.; Bartolomaeus, T.U.P.; Fritsche-Guenther, R.; Rodrigues, A.F.; Yarritu, A.; et al. Quantifying the impact of gut microbiota on inflammation and hypertensive organ damage. Cardiovasc. Res. 2022. [Google Scholar] [CrossRef]
- Joe, B.; McCarthy, C.G.; Edwards, J.M.; Cheng, X.; Chakraborty, S.; Yang, T.; Golonka, R.M.; Mell, B.; Yeo, J.Y.; Bearss, N.R.; et al. Microbiota Introduced to Germ-Free Rats Restores Vascular Contractility and Blood Pressure. Hypertension 2020, 76, 1847–1855. [Google Scholar] [CrossRef]
- Abboud, F.M.; Cicha, M.Z.; Ericsson, A.; Chapleau, M.W.; Singh, M.V. Altering Early Life Gut Microbiota Has Long-Term Effect on Immune System and Hypertension in Spontaneously Hypertensive Rats. Front. Physiol. 2021, 12, 752924. [Google Scholar] [CrossRef]
- Jiang, S.; Shui, Y.; Cui, Y.; Tang, C.; Wang, X.; Qiu, X.; Hu, W.; Fei, L.; Li, Y.; Zhang, S.; et al. Gut microbiota dependent trimethylamine N-oxide aggravates angiotensin II-induced hypertension. Redox Biol. 2021, 46, 102115. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, J.F.; Aden, L.A.; Barbaro, N.R.; Van Beusecum, J.P.; Xiao, L.; Simmons, A.J.; Warden, C.; Pasic, L.; Himmel, L.E.; Washington, M.K.; et al. High dietary salt-induced dendritic cell activation underlies microbial dysbiosis-associated hypertension. JCI Insight 2019, 5, e126241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, S.; Galla, S.; Cheng, X.; Yeo, J.Y.; Mell, B.; Singh, V.; Yeoh, B.; Saha, P.; Mathew, A.V.; Vijay-Kumar, M.; et al. Salt-Responsive Metabolite, beta-Hydroxybutyrate, Attenuates Hypertension. Cell Rep. 2018, 25, 677–689. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.H. Causal association of gut microbiome on the risk of rheumatoid arthritis: A Mendelian randomisation study. Ann. Rheum. Dis. 2022, 81, e3. [Google Scholar] [CrossRef] [Green Version]
- Kishikawa, T.; Maeda, Y.; Nii, T.; Motooka, D.; Matsumoto, Y.; Matsushita, M.; Matsuoka, H.; Yoshimura, M.; Kawada, S.; Teshigawara, S.; et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann. Rheum. Dis. 2020, 79, 103–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Evidence of the Immune Relevance of Prevotella copri, a Gut Microbe, in Patients with Rheumatoid Arthritis. Arthritis Rheumatol. 2017, 69, 964–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhang, S.X.; Yin, X.F.; Zhang, M.X.; Qiao, J.; Xin, X.H.; Chang, M.J.; Gao, C.; Li, Y.F.; Li, X.F. The Gut Microbiota and Its Relevance to Peripheral Lymphocyte Subpopulations and Cytokines in Patients with Rheumatoid Arthritis. J. Immunol. Res. 2021, 2021, 6665563. [Google Scholar] [CrossRef]
- Teng, F.; Klinger, C.N.; Felix, K.M.; Bradley, C.P.; Wu, E.; Tran, N.L.; Umesaki, Y.; Wu, H.J. Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer′s Patch T Follicular Helper Cells. Immunity 2016, 44, 875–888. [Google Scholar] [CrossRef] [Green Version]
- Bates, N.A.; Li, A.; Fan, T.; Cutcliffe, M.P.; Dagenet, C.B.; Sleiman, K.C.; Ma, H.; Tahsin, S.; Garrett, C.S.; Altemus, J.; et al. Gut Commensal Segmented Filamentous Bacteria Fine-Tune T Follicular Regulatory Cells to Modify the Severity of Systemic Autoimmune Arthritis. J. Immunol. 2021, 206, 941–952. [Google Scholar] [CrossRef]
- Teng, F.; Felix, K.M.; Bradley, C.P.; Naskar, D.; Ma, H.; Raslan, W.A.; Wu, H.J. The impact of age and gut microbiota on Th17 and Tfh cells in K/BxN autoimmune arthritis. Arthritis Res. Ther. 2017, 19, 188. [Google Scholar] [CrossRef] [Green Version]
- Block, K.E.; Zheng, Z.; Dent, A.L.; Kee, B.L.; Huang, H. Gut Microbiota Regulates K/BxN Autoimmune Arthritis through Follicular Helper T but Not Th17 Cells. J. Immunol. 2016, 196, 1550–1557. [Google Scholar] [CrossRef] [PubMed]
- Simpkins, D.A.; Downton, P.; Gray, K.J.; Dickson, S.H.; Maidstone, R.J.; Konkel, J.E.; Hepworth, M.R.; Ray, D.W.; Bechtold, D.A.; Gibbs, J.E. Consequences of collagen induced inflammatory arthritis on circadian regulation of the gut microbiome. FASEB J. 2023, 37, e22704. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chu, Y.; Li, J.; Meng, Q.; Liu, Y.; Jin, J.; Wang, Y.; Wang, J.; Huang, B.; Shi, L.; et al. Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis. Sci. Adv. 2022, 8, eabm1511. [Google Scholar] [CrossRef]
- Takahashi, D.; Hoshina, N.; Kabumoto, Y.; Maeda, Y.; Suzuki, A.; Tanabe, H.; Isobe, J.; Yamada, T.; Muroi, K.; Yanagisawa, Y.; et al. Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine 2020, 58, 102913. [Google Scholar] [CrossRef] [PubMed]
- Rosser, E.C.; Piper, C.J.M.; Matei, D.E.; Blair, P.A.; Rendeiro, A.F.; Orford, M.; Alber, D.G.; Krausgruber, T.; Catalan, D.; Klein, N.; et al. Microbiota-Derived Metabolites Suppress Arthritis by Amplifying Aryl-Hydrocarbon Receptor Activation in Regulatory B Cells. Cell Metab. 2020, 31, 837–851. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Guo, Y.; Wang, H.; Yin, A.; Hu, J.; Yuan, T.; Zhou, S.; Xu, W.; Wei, P.; Yin, S.; et al. Gut commensal Parabacteroides distasonis alleviates inflammatory arthritis. Gut 2023. [Google Scholar] [CrossRef]
- Adeyeye, T.E.; Yeung, E.H.; McLain, A.C.; Lin, S.; Lawrence, D.A.; Bell, E.M. Wheeze and Food Allergies in Children Born via Cesarean Delivery: The Upstate KIDS Study. Am. J. Epidemiol. 2019, 188, 355–362. [Google Scholar] [CrossRef]
- Loh, W.; Tang, M.L.K. The Epidemiology of Food Allergy in the Global Context. Int J. Environ. Res. Public Health 2018, 15, 2043. [Google Scholar] [CrossRef] [Green Version]
- Leung, A.S.Y.; Wong, G.W.K.; Tang, M.L.K. Food allergy in the developing world. J. Allergy Clin. Immunol. 2018, 141, 76–78. [Google Scholar] [CrossRef] [Green Version]
- Borbet, T.C.; Pawline, M.B.; Zhang, X.; Wipperman, M.F.; Reuter, S.; Maher, T.; Li, J.; Iizumi, T.; Gao, Z.; Daniele, M.; et al. Influence of the early-life gut microbiota on the immune responses to an inhaled allergen. Mucosal Immunol. 2022, 15, 1000–1011. [Google Scholar] [CrossRef]
- Zhang, Q.; Cheng, L.; Wang, J.; Hao, M.; Che, H. Antibiotic-Induced Gut Microbiota Dysbiosis Damages the Intestinal Barrier, Increasing Food Allergy in Adult Mice. Nutrients 2021, 13, 3315. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Bonilla-Rosso, G.; Kwong Chung, C.K.C.; Bariswyl, L.; Rodriguez, M.P.; Kim, B.S.; Engel, P.; Noti, M. High dietary fat intake induces a microbiota signature that promotes food allergy. J. Allergy Clin. Immunol. 2019, 144, 157–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, P.; Messina, N.; Mohn, W.W.; Finlay, B.B.; Curtis, N. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: A systematic review. J. Allergy Clin. Immunol. 2019, 143, 467–485. [Google Scholar] [CrossRef] [PubMed]
- McCauley, K.E.; Rackaityte, E.; LaMere, B.; Fadrosh, D.W.; Fujimura, K.E.; Panzer, A.R.; Lin, D.L.; Lynch, K.V.; Halkias, J.; Mendoza, V.F.; et al. Heritable vaginal bacteria influence immune tolerance and relate to early-life markers of allergic sensitization in infancy. Cell Rep. Med. 2022, 3, 100713. [Google Scholar] [CrossRef]
- Okba, A.M.; Saber, S.M.; Abdel-Rehim, A.S.; Amin, M.M.; Mohamed, D.A. Fecal microbiota profile in atopic asthmatic adult patients. Eur. Ann. Allergy Clin. Immunol. 2018, 50, 117–124. [Google Scholar] [CrossRef]
- Lundstrom, S.L.; Yang, J.; Kallberg, H.J.; Thunberg, S.; Gafvelin, G.; Haeggstrom, J.Z.; Gronneberg, R.; Grunewald, J.; van Hage, M.; Hammock, B.D.; et al. Allergic asthmatics show divergent lipid mediator profiles from healthy controls both at baseline and following birch pollen provocation. PLoS ONE 2012, 7, e33780. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [Green Version]
- Levan, S.R.; Stamnes, K.A.; Lin, D.L.; Panzer, A.R.; Fukui, E.; McCauley, K.; Fujimura, K.E.; McKean, M.; Ownby, D.R.; Zoratti, E.M.; et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. 2019, 4, 1851–1861. [Google Scholar] [CrossRef]
- Abdulla, O.A.; Neamah, W.; Sultan, M.; Alghetaa, H.K.; Singh, N.; Busbee, P.B.; Nagarkatti, M.; Nagarkatti, P. The Ability of AhR Ligands to Attenuate Delayed Type Hypersensitivity Reaction Is Associated with Alterations in the Gut Microbiota. Front. Immunol. 2021, 12, 684727. [Google Scholar] [CrossRef]
- Roduit, C.; Frei, R.; Ferstl, R.; Loeliger, S.; Westermann, P.; Rhyner, C.; Schiavi, E.; Barcik, W.; Rodriguez-Perez, N.; Wawrzyniak, M.; et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019, 74, 799–809. [Google Scholar] [CrossRef]
- Cait, A.; Hughes, M.R.; Antignano, F.; Cait, J.; Dimitriu, P.A.; Maas, K.R.; Reynolds, L.A.; Hacker, L.; Mohr, J.; Finlay, B.B.; et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 2018, 11, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Thorburn, A.N.; McKenzie, C.I.; Shen, S.; Stanley, D.; Macia, L.; Mason, L.J.; Roberts, L.K.; Wong, C.H.; Shim, R.; Robert, R.; et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015, 6, 7320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.; McKenzie, C.; Vuillermin, P.J.; Goverse, G.; Vinuesa, C.G.; Mebius, R.E.; Macia, L.; Mackay, C.R. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep. 2016, 15, 2809–2824. [Google Scholar] [CrossRef] [Green Version]
- Arifuzzaman, M.; Won, T.H.; Li, T.T.; Yano, H.; Digumarthi, S.; Heras, A.F.; Zhang, W.; Parkhurst, C.N.; Kashyap, S.; Jin, W.B.; et al. Inulin fibre promotes microbiota-derived bile acids and type 2 inflammation. Nature 2022, 611, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Seki, D.; Mayer, M.; Hausmann, B.; Pjevac, P.; Giordano, V.; Goeral, K.; Unterasinger, L.; Klebermass-Schrehof, K.; De Paepe, K.; Van de Wiele, T.; et al. Aberrant gut-microbiota-immune-brain axis development in premature neonates with brain damage. Cell Host Microbe 2021, 29, 1558–1572. [Google Scholar] [CrossRef]
- Ouabbou, S.; He, Y.; Butler, K.; Tsuang, M. Inflammation in Mental Disorders: Is the Microbiota the Missing Link? Neurosci. Bull. 2020, 36, 1071–1084. [Google Scholar] [CrossRef]
- Slyepchenko, A.; Carvalho, A.F.; Cha, D.S.; Kasper, S.; McIntyre, R.S. Gut emotions—Mechanisms of action of probiotics as novel therapeutic targets for depression and anxiety disorders. CNS Neurol. Disord. Drug Targets 2014, 13, 1770–1786. [Google Scholar] [CrossRef]
- Zhang, X.; Hou, Y.; Li, Y.; Wei, W.; Cai, X.; Shao, H.; Yuan, Y.; Zheng, X. Taxonomic and Metabolic Signatures of Gut Microbiota for Assessing the Severity of Depression and Anxiety in Major Depressive Disorder Patients. Neuroscience 2022, 496, 179–189. [Google Scholar] [CrossRef]
- Galley, J.D.; Mashburn-Warren, L.; Blalock, L.C.; Lauber, C.L.; Carroll, J.E.; Ross, K.M.; Hobel, C.; Coussons-Read, M.; Dunkel Schetter, C.; Gur, T.L. Maternal anxiety, depression and stress affects offspring gut microbiome diversity and bifidobacterial abundances. Brain Behav. Immun. 2023, 107, 253–264. [Google Scholar] [CrossRef]
- O′Neill, S.M.; Curran, E.A.; Dalman, C.; Kenny, L.C.; Kearney, P.M.; Clarke, G.; Cryan, J.F.; Dinan, T.G.; Khashan, A.S. Birth by Caesarean Section and the Risk of Adult Psychosis: A Population-Based Cohort Study. Schizophr. Bull. 2016, 42, 633–641. [Google Scholar] [CrossRef] [Green Version]
- Morais, L.H.; Golubeva, A.V.; Casey, S.; Scott, K.A.; Ramos Costa, A.P.; Moloney, G.M.; Dinan, T.G.; Cryan, J.F. Early-life oxytocin attenuates the social deficits induced by caesarean-section delivery in the mouse. Neuropsychopharmacology 2021, 46, 1958–1968. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.M.K.; Cowan, C.S.M.; Bastiaanssen, T.F.S.; Moloney, G.M.; Theune, N.; van de Wouw, M.; Florensa Zanuy, E.; Ventura-Silva, A.P.; Codagnone, M.G.; Villalobos-Manriquez, F.; et al. Critical windows of early-life microbiota disruption on behaviour, neuroimmune function, and neurodevelopment. Brain Behav. Immun. 2022, 108, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pu, F.; Peng, C.; Wang, Y.; Zhang, Y.; Wu, S.; Wang, S.; Shen, X.; Li, Y.; Cheng, R.; et al. Antibiotic cocktail-induced gut microbiota depletion in different stages could cause host cognitive impairment and emotional disorders in adulthood in different manners. Neurobiol. Dis. 2022, 170, 105757. [Google Scholar] [CrossRef] [PubMed]
- Cuskelly, A.; Hoedt, E.C.; Harms, L.; Talley, N.J.; Tadros, M.A.; Keely, S.; Hodgson, D.M. Neonatal immune challenge influences the microbiota and behaviour in a sexually dimorphic manner. Brain Behav. Immun. 2022, 103, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Venna, V.R.; Durgan, D.J.; Shi, H.; Hudobenko, J.; Putluri, N.; Petrosino, J.; McCullough, L.D.; Bryan, R.M. Young versus aged microbiota transplants to germ-free mice: Increased short-chain fatty acids and improved cognitive performance. Gut Microbes 2020, 12, 1814107. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, I.; Gao, I.H.; Lin, W.Y.; Allen, M.; Li, X.V.; Fiers, W.D.; De Celie, M.B.; Putzel, G.G.; Yantiss, R.K.; Johncilla, M.; et al. Mucosal fungi promote gut barrier function and social behavior via Type 17 immunity. Cell 2022, 185, 831–846. [Google Scholar] [CrossRef]
- Allahverdiyev, A.; Tari, G.; Bagirova, M.; Abamor, E.S. Current Approaches in Development of Immunotherapeutic Vaccines for Breast Cancer. J. Breast Cancer 2018, 21, 343–353. [Google Scholar] [CrossRef]
- Johnson, L.A.; June, C.H. Driving gene-engineered T cell immunotherapy of cancer. Cell Res. 2017, 27, 38–58. [Google Scholar] [CrossRef] [Green Version]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [Green Version]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Wang, T.; Tu, X.; Huang, Y.; Zhang, H.; Tan, D.; Jiang, W.; Cai, S.; Zhao, P.; Song, R.; et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J. Immunother. Cancer 2019, 7, 193. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Dong, H.; Xia, L.; Yang, Y.; Zhu, Y.; Shen, Y.; Zheng, H.; Yao, C.; Wang, Y.; Lu, S. The Diversity of Gut Microbiome is Associated with Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J. Thorac. Oncol. 2019, 14, 1378–1389. [Google Scholar] [CrossRef]
- Li, L.; Ye, J. Characterization of gut microbiota in patients with primary hepatocellular carcinoma received immune checkpoint inhibitors: A Chinese population-based study. Medicine 2020, 99, e21788. [Google Scholar] [CrossRef]
- Coutzac, C.; Jouniaux, J.M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef]
- Zhang, S.L.; Han, B.; Mao, Y.Q.; Zhang, Z.Y.; Li, Z.M.; Kong, C.Y.; Wu, Y.; Chen, G.Q.; Wang, L.S. Lacticaseibacillus paracasei sh2020 induced antitumor immunity and synergized with anti-programmed cell death 1 to reduce tumor burden in mice. Gut Microbes 2022, 14, 2046246. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Wang, Y.; Chen, X.; Wang, C.; Chen, X.; Yuan, X.; Liu, L.; Yang, J.; Zhou, X. Prevotellaceae produces butyrate to alleviate PD-1/PD-L1 inhibitor-related cardiotoxicity via PPARalpha-CYP4X1 axis in colonic macrophages. J. Exp. Clin. Cancer Res. 2022, 41, 1. [Google Scholar] [CrossRef]
- Raad, M.A.; Chams, N.H.; Sharara, A.I. New and Evolving Immunotherapy in Inflammatory Bowel Disease. Inflamm. Intest. Dis. 2016, 1, 85–95. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Johnson, J.R. Medical and economic impact of extraintestinal infections due to Escherichia coli: Focus on an increasingly important endemic problem. Microbes Infect. 2003, 5, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Collotta, D.; Hull, W.; Mastrocola, R.; Chiazza, F.; Cento, A.S.; Murphy, C.; Verta, R.; Alves, G.F.; Gaudioso, G.; Fava, F.; et al. Baricitinib counteracts metaflammation, thus protecting against diet-induced metabolic abnormalities in mice. Mol. Metab. 2020, 39, 101009. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ding, X.; Liu, Y.; Marcella, C.; Dai, M.; Zhang, T.; Bai, J.; Xiang, L.; Wen, Q.; Cui, B.; et al. Fecal Microbiota Transplantation is a Promising Switch Therapy for Patients with Prior Failure of Infliximab in Crohn′s Disease. Front. Pharmacol. 2021, 12, 658087. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Dong, F.; Li, X.; Li, Y.; Yu, G.; Liu, Z.; Wang, Y.; Zhang, T. Bifidobacterium longum CECT 7894 Improves the Efficacy of Infliximab for DSS-Induced Colitis via Regulating the Gut Microbiota and Bile Acid Metabolism. Front. Pharmacol. 2022, 13, 902337. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.J.; Plichta, D.; Hogstrom, L.; Borren, N.Z.; Lau, H.; Gregory, S.M.; Tan, W.; Khalili, H.; Clish, C.; Vlamakis, H.; et al. Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease. Cell Host Microbe 2021, 29, 1294–1304. [Google Scholar] [CrossRef] [PubMed]
- Sanchis-Artero, L.; Martinez-Blanch, J.F.; Manresa-Vera, S.; Cortes-Castell, E.; Valls-Gandia, M.; Iborra, M.; Paredes-Arquiola, J.M.; Bosca-Watts, M.; Huguet, J.M.; Gil-Borras, R.; et al. Evaluation of changes in intestinal microbiota in Crohn′s disease patients after anti-TNF alpha treatment. Sci. Rep. 2021, 11, 10016. [Google Scholar] [CrossRef]
- Ding, N.S.; McDonald, J.A.K.; Perdones-Montero, A.; Rees, D.N.; Adegbola, S.O.; Misra, R.; Hendy, P.; Penez, L.; Marchesi, J.R.; Holmes, E.; et al. Metabonomics and the Gut Microbiome Associated with Primary Response to Anti-TNF Therapy in Crohn′s Disease. J. Crohn’s Colitis 2020, 14, 1090–1102. [Google Scholar] [CrossRef]
- Serfaty, L.; De Leusse, A.; Rosmorduc, O.; Desaint, B.; Flejou, J.F.; Chazouilleres, O.; Poupon, R.E.; Poupon, R. Ursodeoxycholic acid therapy and the risk of colorectal adenoma in patients with primary biliary cirrhosis: An observational study. Hepatology 2003, 38, 203–209. [Google Scholar] [CrossRef]
- Shah, S.A.; Volkov, Y.; Arfin, Q.; Abdel-Latif, M.M.; Kelleher, D. Ursodeoxycholic acid inhibits interleukin 1 beta [corrected] and deoxycholic acid-induced activation of NF-kappaB and AP-1 in human colon cancer cells. Int. J. Cancer 2006, 118, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Lu, C.; Song, Z.; Qiao, C.; Wang, J.; Chen, J.; Zhang, C.; Zeng, X.; Ma, Z.; Chen, T.; et al. Ursodeoxycholic acid reduces antitumor immunosuppression by inducing CHIP-mediated TGF-beta degradation. Nat. Commun. 2022, 13, 3419. [Google Scholar] [CrossRef] [PubMed]
- Grover, M.; Kashyap, P.C. Germ-free mice as a model to study effect of gut microbiota on host physiology. Neurogastroenterol. Motil. 2014, 26, 745–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coyte, K.Z.; Rakoff-Nahoum, S. Understanding Competition and Cooperation within the Mammalian Gut Microbiome. Curr. Biol. 2019, 29, R538–R544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehtimaki, J.; Thorsen, J.; Rasmussen, M.A.; Hjelmso, M.; Shah, S.; Mortensen, M.S.; Trivedi, U.; Vestergaard, G.; Bonnelykke, K.; Chawes, B.L.; et al. Urbanized microbiota in infants, immune constitution, and later risk of atopic diseases. J. Allergy Clin. Immunol. 2021, 148, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Arnesen, H.; Hitch, T.C.A.; Steppeler, C.; Muller, M.H.B.; Knutsen, L.E.; Gunnes, G.; Angell, I.L.; Ormaasen, I.; Rudi, K.; Paulsen, J.E.; et al. Naturalizing laboratory mice by housing in a farmyard-type habitat confers protection against colorectal carcinogenesis. Gut Microbes 2021, 13, 1993581. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [Green Version]
- Aguiar-Pulido, V.; Huang, W.; Suarez-Ulloa, V.; Cickovski, T.; Mathee, K.; Narasimhan, G. Metagenomics, Metatranscriptomics, and Metabolomics Approaches for Microbiome Analysis. Evol. Bioinform. Online 2016, 12, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Dias, C.K.; Starke, R.; Pylro, V.S.; Morais, D.K. Database limitations for studying the human gut microbiome. PeerJ Comput. Sci. 2020, 6, e289. [Google Scholar] [CrossRef]
- Porras, A.M.; Shi, Q.; Zhou, H.; Callahan, R.; Montenegro-Bethancourt, G.; Solomons, N.; Brito, I.L. Geographic differences in gut microbiota composition impact susceptibility to enteric infection. Cell Rep. 2021, 36, 109457. [Google Scholar] [CrossRef]
- Long, L.L.; Svenson, K.L.; Mourino, A.J.; Michaud, M.; Fahey, J.R.; Waterman, L.; Vandegrift, K.L.; Adams, M.D. Shared and distinctive features of the gut microbiome of C57BL/6 mice from different vendors and production sites, and in response to a new vivarium. Lab. Anim. 2021, 50, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]



| Diseases | Reference | Findings |
|---|---|---|
| Gastrointestinal Infections | Singer et al., 2019 [222] | Provide resistance against colonization and invasion by pathobiont. |
| Tovaglieri et al., 2019 [223] | Human gut microbiome metabolites induce expression of flagellin (a bacterial protein) increases EHEC motility and epithelial injury. | |
| IBD | Lee and chang, 2021 [224] | Gut microbiota dysbiosis of IBD patients is consistently marked by an overgrowth in Proteobacteria. |
| Furusawa et al., 2013 [110] | SCFA confers protection against IBD by maintaining gut barrier integrity, promoting Treg cell differentiation, and inhibiting histone deacetylases. | |
| Colorectal carcinoma | Sepich-Poore et al., 2021 [225] | Generation of genotoxin such as Bacteroides fragilis toxin (Bft), cytolethal distending toxin (CDT), and colibactin. |
| Hale et al., 2017 [226] | Bacterial-derived secondary bile acids and hydrogen sulfide promote proinflammatory milieu that increases CRC risk. | |
| Yeoh et al., 2020 [227] | Bacteria such as F. nucleatum can adhere to colon tumors and aggravate tumorigenesis. | |
| Hepatocellular carcinoma | Lin et al., 1995 [228] | Systemic translocation of LPS promotes chronic liver injury and predisposes to HCC. |
| Singh et al., 2018 [229] | Excess butyrate production promotes HCC progression. | |
| Yoshimoto et al., 2013 [230] | Secondary bile acids promote carcinogenesis and impede anti-tumor immunosurveillance in the liver. | |
| Cardiometabolic disease | Cani et al., 2007 [231] Guasch-Ferré et al., 2017 [232] Millard et al., 2018 [233] | LPS and other microbial ligands drive low-grade chronic inflammation and predispose to CVD. |
| Bacterial trimethylamine and its conversion to trimethylamine-N-oxide in the liver increases the risk of coronary artery disease, metabolic syndrome, stroke, and vascular inflammation. | ||
| Rheumatoid Arthritis | Scher et al., 2013 [234] | Prevotella spp. Abundance is positively associated with new-onset rheumatoid arthritis. |
| Allergic Diseases | Fazlollahi et al., 2018 [235] Bunyavanich et al., 2016 [236] | Gut microbiota dysbiosis increases risk for allergic disease, e.g., food allergy and asthma. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campbell, C.; Kandalgaonkar, M.R.; Golonka, R.M.; Yeoh, B.S.; Vijay-Kumar, M.; Saha, P. Crosstalk between Gut Microbiota and Host Immunity: Impact on Inflammation and Immunotherapy. Biomedicines 2023, 11, 294. https://doi.org/10.3390/biomedicines11020294
Campbell C, Kandalgaonkar MR, Golonka RM, Yeoh BS, Vijay-Kumar M, Saha P. Crosstalk between Gut Microbiota and Host Immunity: Impact on Inflammation and Immunotherapy. Biomedicines. 2023; 11(2):294. https://doi.org/10.3390/biomedicines11020294
Chicago/Turabian StyleCampbell, Connor, Mrunmayee R. Kandalgaonkar, Rachel M. Golonka, Beng San Yeoh, Matam Vijay-Kumar, and Piu Saha. 2023. "Crosstalk between Gut Microbiota and Host Immunity: Impact on Inflammation and Immunotherapy" Biomedicines 11, no. 2: 294. https://doi.org/10.3390/biomedicines11020294
APA StyleCampbell, C., Kandalgaonkar, M. R., Golonka, R. M., Yeoh, B. S., Vijay-Kumar, M., & Saha, P. (2023). Crosstalk between Gut Microbiota and Host Immunity: Impact on Inflammation and Immunotherapy. Biomedicines, 11(2), 294. https://doi.org/10.3390/biomedicines11020294