A Simple and Affordable Method to Create Nonsense Mutation Clones of p53 for Studying the Premature Termination Codon Readthrough Activity of PTC124
Abstract
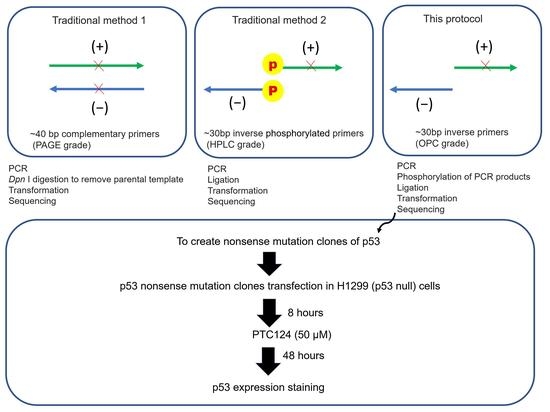
:1. Introduction
2. Experimental Design
2.1. Materials
- OPC purification primers (Genomics, New Taipei City, Taiwan) (mutated site is underlined)p53 W91X: (−) 5′-ACAGGGGTCAGGAGGGGGCT-3′ and(+) 5′-CATCTTCTGTCCCTTCCCAGAAAACCTACCA-3′p53 S94X: (−) 5′-GGGACAGAAGATCACAGGGGCCAGG-3′ and(+) 5′-TTCCCAGAAAACCTACCAGGGCAGCTAC-3′p53 R306X: (−) 5′-GGGCAGTGCTCACTTAGTGCTCCCT-3′ and(+) 5′-AACAACACCAGCTCCTCTCCCCAGC-3′p53 R342X: (−) 5′-GAAGCGCTCACGCCCACGGAT-3′ and(+) 5′-GAGATGTTCTGAGAGCTGAATGAGGCCTTGGAA-3′
- CloneAmp HiFi PCR Premix (TaKaRa Bio, Shiga, Japan; #639298)
- Phusion Plus PCR Master Mix (Thermo Scientific, Waltham, MA, USA; #F631S)
- Phusion High-Fidelity PCR Master Mix (Thermo Scientific, Waltham, MA, USA; #F-531S)
- T4 polynucleotide kinase (T4 PNK) (New England Biolabs, Hitchin, UK; #M0201S)
- YB Rapid Ligation Kit (Yeastern Biotech, Taipei City, Taiwan; # FYC003-100R); each kit contains yT4 DNA Ligase 100 μL (3 U/μL), 10× Ligation Buffer A, and 10× Ligation Buffer B.
- ECOS 9-5 Competent Cells [strain JM109] (Yeastern Biotech, Taipei City, Taiwan; #FYE707-10VL)
- GeneJET Plasmid Midiprep Kit (Thermo Scientific, Waltham, MA, USA; #K0481)
- Opt-MEM medium (Thermo Scientific, Waltham, MA, USA; #31985062)
- TransIT-X2 Transfection Reagent (Mirus Bio, Madison, WI, USA; #MIR6000)
- PTC124 (MedChemExpress, Monmouth Junction, NJ, USA; #HY-14832)
- Primary antibody p53 (clone DO-1; Santa Cruz, Dallas, TX, USA; #sc-126) recognized N-terminal epitope mapping amino acid residues 11–25 of human p53.
- Primary antibody p53 (clone PAb 122; Thermo Scientific, Waltham, MA, USA; #MA5-12453), recognizes C-terminal epitope mapping amino acid residues 370–378 of human p53.
- Secondary antibody PE-conjugated mouse IgG (Thermo Scientific, Waltham, MA, USA; #M30004-1)
- Hoechst 33342 (Tocris Bioscience, Ellisville, MS, USA; #5117) was prepared as stock solution (1 mg/mL in water)
2.2. Equipment
- SimpliAmp Thermal Cycler (Applied Biosystems, Waltham, MA, USA; #A24811)
- NanoDrop ND-1000 (Thermo Scientific, Waltham, MA, USA; #ND1000WOC)
- ECLIPSE Ts2 inverted fluorescence microscope (Nikon, Tokyo, Japan; #094604D)
3. Procedure
3.1. Part 1: Creation of p53 Nonsense Mutation Clones (Modified Inverse PCR-Based Site-Directed Mutagenesis)
3.1.1. Inverse PCR
- CloneAmp HiFi PCR Premix or Phusion High-Fidelity PCR Master Mix (Thermo Scientific, Waltham, MA, USA; #F-531S) or Phusion High-Fidelity PCR Master Mix: 10 µL
- Upstream primer (−) 5 µM: 1 µL
- Downstream primer (+) 5 µM: 1 µL
- p53 wild-type plasmid (pcDNA 3.0 p53) 1 ng/µL: 1 µL
- Sterilized distilled water: 7 µL
- Initial denaturation: 98 °C for 30 s
- Denaturation: 98 °C for 10 s
- Annealing and extension: 72 °C for 2 min
- Final extension: 72 °C for 5 min
3.1.2. T4 PNK Reaction
3.1.3. Ligation
3.1.4. Transformation
3.1.5. Sequencing
3.2. Part 2: Transfection of p53 Nonsense Mutation Clones into p53 Null Cells
3.2.1. Plasmid Isolation
3.2.2. Liposome-Mediated Transfection
3.3. Part 3: Adding PTC124 and Determining p53 Expression
4. Results
5. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar] [CrossRef]
- Hug, N.; Longman, D.; Cáceres, J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016, 44, 1483–1495. [Google Scholar] [CrossRef]
- Floquet, C.; Deforges, J.; Rousset, J.P.; Bidou, L. Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 2011, 39, 3350–3362. [Google Scholar] [CrossRef]
- Zhang, M.; Heldin, A.; Palomar-Siles, M.; Öhlin, S.; Bykov, V.J.N.; Wiman, K.G. Synergistic Rescue of Nonsense Mutant Tumor Suppressor p53 by Combination Treatment with Aminoglycosides and Mdm2 Inhibitors. Front. Oncol. 2017, 7, 323. [Google Scholar] [CrossRef] [PubMed]
- Auld, D.S.; Thorne, N.; Maguire, W.F.; Inglese, J. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proc. Natl. Acad. Sci. USA 2009, 106, 3585–3590. [Google Scholar] [CrossRef] [PubMed]
- Bezzerri, V.; Lentini, L.; Api, M.; Busilacchi, E.M.; Cavalieri, V.; Pomilio, A.; Diomede, F.; Pegoraro, A.; Cesaro, S.; Poloni, A.; et al. Novel Translational Read-through-Inducing Drugs as a Therapeutic Option for Shwachman-Diamond Syndrome. Biomedicines 2022, 10, 886. [Google Scholar] [CrossRef]
- Rybak, L.P.; Kelly, T. Ototoxicity: Bioprotective mechanisms. Curr. Opin. Otolaryngol. Head Neck Surg. 2003, 11, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Selimoglu, E. Aminoglycoside-induced ototoxicity. Curr. Pharm. Des. 2007, 13, 119–126. [Google Scholar] [CrossRef]
- Kandasamy, J.; Atia-Glikin, D.; Shulman, E.; Shapira, K.; Shavit, M.; Belakhov, V.; Baasov, T. Increased selectivity toward cytoplasmic versus mitochondrial ribosome confers improved efficiency of synthetic aminoglycosides in fixing damaged genes: A strategy for treatment of genetic diseases caused by nonsense mutations. J. Med. Chem. 2012, 55, 10630–10643. [Google Scholar] [CrossRef]
- Finkel, R.S.; Flanigan, K.M.; Wong, B.; Bönnemann, C.; Sampson, J.; Sweeney, H.L.; Reha, A.; Northcutt, V.J.; Elfring, G.; Barth, J.; et al. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS ONE 2013, 8, e81302. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Ginjaar, I.B.; Bushby, K. The importance of genetic diagnosis for Duchenne muscular dystrophy. J. Med. Genet. 2016, 53, 145–151. [Google Scholar] [CrossRef]
- Cipolli, M.; Borgatti, M.; Poloni, A.; Bronte, V.; Fabrizzi, B.; Sorio, C.; Gambari, R.; D’Amico, G.; Marinelli Busilacchi, E.; Allegri, M.; et al. Breakthroughs in Preclinical Development of Ataluren (PTC124) As Therapeutic Option for Patients Affected By Shwachman-Diamond Syndrome: Towards the First Clinical Trial. Blood 2019, 134, 451. [Google Scholar] [CrossRef]
- Burroughs, L.; Woolfrey, A.; Shimamura, A. Shwachman-Diamond syndrome: A review of the clinical presentation, molecular pathogenesis, diagnosis, and treatment. Hematol. Oncol. Clin. N. Am. 2009, 23, 233–248. [Google Scholar] [CrossRef]
- Bezzerri, V.; Bardelli, D.; Morini, J.; Vella, A.; Cesaro, S.; Sorio, C.; Biondi, A.; Danesino, C.; Farruggia, P.; Assael, B.M.; et al. Ataluren-driven restoration of Shwachman-Bodian-Diamond syndrome protein function in Shwachman-Diamond syndrome bone marrow cells. Am. J. Hematol. 2018, 93, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, F.; Cao, Y.; Xu, H.; Wu, Y.; Wu, S.; Liu, D.; Zhao, Y.; Songyang, Z.; Ma, W. Shwachman-Diamond Syndrome Protein SBDS Maintains Human Telomeres by Regulating Telomerase Recruitment. Cell Rep. 2018, 22, 1849–1860. [Google Scholar] [CrossRef]
- Wu, M.H.; Lu, R.Y.; Yu, S.J.; Tsai, Y.Z.; Lin, Y.C.; Bai, Z.Y.; Liao, R.Y.; Hsu, Y.C.; Chen, C.C.; Cai, B.H. PTC124 Rescues Nonsense Mutation of Two Tumor Suppressor Genes NOTCH1 and FAT1 to Repress HNSCC Cell Proliferation. Biomedicines 2022, 10, 2948. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.H.; Chao, C.F.; Huang, H.C.; Lee, H.Y.; Kannagi, R.; Chen, J.Y. Roles of p53 Family Structure and Function in Non-Canonical Response Element Binding and Activation. Int. J. Mol. Sci. 2019, 20, 3681. [Google Scholar] [CrossRef]
- Meng, P.; Zhang, Y.F.; Zhang, W.; Chen, X.; Xu, T.; Hu, S.; Liang, X.; Feng, M.; Yang, X.; Ho, M. Identification of the atypical cadherin FAT1 as a novel glypican-3 interacting protein in liver cancer cells. Sci. Rep. 2021, 11, 40. [Google Scholar] [CrossRef]
- Platonova, N.; Manzo, T.; Mirandola, L.; Colombo, M.; Calzavara, E.; Vigolo, E.; Cermisoni, G.C.; De Simone, D.; Garavelli, S.; Cecchinato, V.; et al. PI3K/AKT signaling inhibits NOTCH1 lysosome-mediated degradation. Genes Chromosomes Cancer 2015, 54, 516–526. [Google Scholar] [CrossRef]
- Scott, S.P.; Teh, A.; Peng, C.; Lavin, M.F. One-step site-directed mutagenesis of ATM cDNA in large (20kb) plasmid constructs. Hum. Mutat. 2002, 20, 323. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Cabré, A.; Girona, J.; Zalba, G.; Moreno, M.U.; Díez, J.; Masana, L. Generation of eight adjacent mutations in a single step using a site-directed mutagenesis kit. Clin. Chem. Lab. Med. 2004, 42, 384–386. [Google Scholar] [CrossRef] [PubMed]
- Spiliotis, M. Inverse fusion PCR cloning. PLoS ONE 2012, 7, e35407. [Google Scholar] [CrossRef] [PubMed]
- Andrus, A.; Kuimelis, R.G. Cartridge methods for oligonucleotide purification. Curr. Protoc. Nucleic Acid Chem. 2001, 1, 10.7.1–10.7.5. [Google Scholar] [CrossRef]
- Boom, R.; Sol, C.J.; Salimans, M.M.; Jansen, C.L.; Wertheim-van Dillen, P.M.; van der Noordaa, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Studzińska, S.; Nuckowski, Ł.; Buszewski, B. Oligonucleotides Isolation and Separation-A Review on Adsorbent Selection. Int. J. Mol. Sci. 2022, 23, 9546. [Google Scholar] [CrossRef]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S.; et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef]
- Balijepalli, S.Y.; Anderson, C.L.; Lin, E.C.; January, C.T. Rescue of mutated cardiac ion channels in inherited arrhythmia syndromes. J. Cardiovasc. Pharmacol. 2010, 56, 113–122. [Google Scholar] [CrossRef]
- Yu, H.; Liu, X.; Huang, J.; Zhang, Y.; Hu, R.; Pu, J. Comparison of read-through effects of aminoglycosides and PTC124 on rescuing nonsense mutations of HERG gene associated with long QT syndrome. Int. J. Mol. Med. 2014, 33, 729–735. [Google Scholar] [CrossRef]
- Azimov, R.; Abuladze, N.; Sassani, P.; Newman, D.; Kao, L.; Liu, W.; Orozco, N.; Ruchala, P.; Pushkin, A.; Kurtz, I. G418-mediated ribosomal read-through of a nonsense mutation causing autosomal recessive proximal renal tubular acidosis. Am. J. Physiol. Ren. Physiol. 2008, 295, F633–F641. [Google Scholar] [CrossRef]
- Friesen, W.J.; Trotta, C.R.; Tomizawa, Y.; Zhuo, J.; Johnson, B.; Sierra, J.; Roy, B.; Weetall, M.; Hedrick, J.; Sheedy, J.; et al. The nucleoside analog clitocine is a potent and efficacious readthrough agent. RNA 2017, 23, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Trzaska, C.; Amand, S.; Bailly, C.; Leroy, C.; Marchand, V.; Duvernois-Berthet, E.; Saliou, J.M.; Benhabiles, H.; Werkmeister, E.; Chassat, T.; et al. 2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations. Nat. Commun. 2020, 11, 1509. [Google Scholar] [CrossRef] [PubMed]






| Nonsense Mutation Gene | Protein Mutation Site | DNA Mutation Site | Original Codon | Mutated Pre-Stop Codon | Lung Cancer Samples/All Type Samples | COSMIC ID |
|---|---|---|---|---|---|---|
| P53 | p.W91X | c.273G > A | TGG | TGA | 13/69 (23%) | COSM44492 |
| P53 | p.S94X | c.281C > G | TCA | TGA | 1/11 (9%) | COSM45653 |
| P53 | p.R306X | c.916C > T | CGA | TGA | 6/376 (2%) | COSM10663 |
| P53 | p.R342X | c.1024C > T | CGA | TGA | 22/453 (5%) | COSM11073 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.-C.; Liao, R.-Y.; Yeh, F.-Y.; Lin, Y.-R.; Wu, T.-Y.; Pastor, A.E.; Zul, D.D.; Hsu, Y.-C.; Wu, K.-Y.; Liu, K.-F.; et al. A Simple and Affordable Method to Create Nonsense Mutation Clones of p53 for Studying the Premature Termination Codon Readthrough Activity of PTC124. Biomedicines 2023, 11, 1310. https://doi.org/10.3390/biomedicines11051310
Chen C-C, Liao R-Y, Yeh F-Y, Lin Y-R, Wu T-Y, Pastor AE, Zul DD, Hsu Y-C, Wu K-Y, Liu K-F, et al. A Simple and Affordable Method to Create Nonsense Mutation Clones of p53 for Studying the Premature Termination Codon Readthrough Activity of PTC124. Biomedicines. 2023; 11(5):1310. https://doi.org/10.3390/biomedicines11051310
Chicago/Turabian StyleChen, Chia-Chi, Ruo-Yu Liao, Fang-Yu Yeh, Yu-Rou Lin, Tze-You Wu, Alexa Escobar Pastor, Danny Danilo Zul, Yun-Chien Hsu, Kuan-Yo Wu, Ke-Fang Liu, and et al. 2023. "A Simple and Affordable Method to Create Nonsense Mutation Clones of p53 for Studying the Premature Termination Codon Readthrough Activity of PTC124" Biomedicines 11, no. 5: 1310. https://doi.org/10.3390/biomedicines11051310
APA StyleChen, C. -C., Liao, R. -Y., Yeh, F. -Y., Lin, Y. -R., Wu, T. -Y., Pastor, A. E., Zul, D. D., Hsu, Y. -C., Wu, K. -Y., Liu, K. -F., Kannagi, R., Chen, J. -Y., & Cai, B. -H. (2023). A Simple and Affordable Method to Create Nonsense Mutation Clones of p53 for Studying the Premature Termination Codon Readthrough Activity of PTC124. Biomedicines, 11(5), 1310. https://doi.org/10.3390/biomedicines11051310