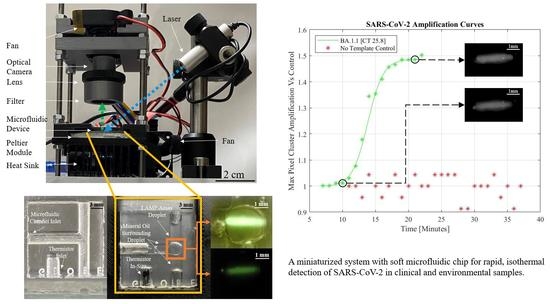
A Miniaturized System for Rapid, Isothermal Detection of SARS-CoV-2 in Human and Environmental Samples
Abstract
:1. Introduction
2. Materials
2.1. Microfluidic Chip Design and Fabrication
2.2. Optical Assembly
2.3. System-on-Board Automation for Temperature Control and Sensor Read-Out
3. Methods
3.1. SARS-CoV-2 Clinical Samples: PCR and Whole Genome Sequencing
3.2. LAMP Primer Design
3.3. Experimental Procedure
3.4. Real-Time Detection and Image Analysis
4. Results and Discussion
4.1. Real-Time Detection of Clinical and Environmental Samples
4.2. Overall Performance of LAMP Based Miniaturized Platform
4.3. Current Capabilities and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [Green Version]
- Zhao, V.X.T.; Wong, T.I.; Zheng, X.T.; Tan, Y.N.; Zhou, X. Colorimetric biosensors for point-of-care virus detections. Mater. Sci. Energy Technol. 2019, 3, 237–249. [Google Scholar] [CrossRef]
- Li, Q.; Yue, Z.; Liu, H.; Liang, C.; Zheng, X.; Zhao, Y.; Chen, X.; Xiao, X.; Chen, C. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of lymphocystis disease virus. J. Virol. Methods 2010, 163, 378–384. [Google Scholar] [CrossRef]
- Hayasaka, D.; Aoki, K.; Morita, K. Development of simple and rapid assay to detect viral RNA of tick-borne encephalitis virus by reverse transcription-loop-mediated isothermal amplification. Virol. J. 2013, 10, 68. [Google Scholar] [CrossRef] [Green Version]
- Mannier, C.; Yoon, J.-Y. Progression of LAMP as a Result of the COVID-19 Pandemic: Is PCR Finally Rivaled? Biosensors 2022, 12, 492. [Google Scholar] [CrossRef]
- Sreejith, K.R.; Umer, M.; Dirr, L.; Bailly, B.; Guillon, P.; von Itzstein, M.; Soda, N.; Kasetsirikul, S.; Shiddiky, M.J.A.; Nguyen, N.-T.; et al. A Portable Device for LAMP Based Detection of SARS-CoV-2. Micromachines 2021, 12, 1151. [Google Scholar] [CrossRef]
- Teymouri, M.; Mollazadeh, S.; Mortazavi, H.; Ghale-Noie, Z.N.; Keyvani, V.; Aghababaei, F.; Hamblin, M.R.; Abbaszadeh-Goudarzi, G.; Pourghadamyari, H.; Hashemian, S.M.R.; et al. Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19. Pathol. Res. Pract. 2021, 221, 153443. [Google Scholar] [CrossRef]
- Liu, H.; Chang, S.; Chen, S.; Du, Y.; Wang, H.; Wang, C.; Xiang, Y.; Wang, Q.; Li, Z.; Wang, S.; et al. Highly sensitive and rapid detection of SARS-CoV-2 via a portable CRISPR-Cas13a-based lateral flow assay. J. Med. Virol. 2022, 94, 5858–5866. [Google Scholar] [CrossRef]
- Van Dongen, J.E.; Berendsen, J.T.; Steenbergen, R.D.; Wolthuis, R.; Eijkel, J.C.; Segerink, L.I. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens. Bioelectron. 2020, 166, 112445. [Google Scholar] [CrossRef]
- Bai, Y.; Ji, J.; Ji, F.; Wu, S.; Tian, Y.; Jin, B.; Li, Z. Recombinase polymerase amplification integrated with microfluidics for nucleic acid testing at point of care. Talanta 2022, 240, 123209. [Google Scholar] [CrossRef]
- Silva, F.S.R.; Erdogmus, E.; Shokr, A.; Kandula, H.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Hardie, J.M.; Pacheco, L.G.C.; Li, J.Z.; Kuritzkes, D.R.; et al. SARS-CoV-2 RNA Detection by a Cellphone-Based Amplification-Free System with CRISPR/CAS-Dependent Enzymatic (CASCADE) Assay. Adv. Mater. Technol. 2021, 6, 2100602. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, X.; Wang, D.; Jiang, X. Recent advancements in nucleic acid detection with microfluidic chip for molecular diagnostics. TrAC Trends Anal. Chem. 2023, 158, 116871. [Google Scholar] [CrossRef]
- Bhardwaj, T.; Ramana, L.N.; Sharma, T.K. Current Advancements and Future Road Map to Develop ASSURED Microfluidic Biosensors for Infectious and Non-Infectious Diseases. Biosensors 2022, 12, 357. [Google Scholar] [CrossRef]
- Deng, H.; Jayawardena, A.; Chan, J.; Tan, S.M.; Alan, T.; Kwan, P. An ultra-portable, self-contained point-of-care nucleic acid amplification test for diagnosis of active COVID-19 infection. Sci. Rep. 2021, 11, 15176. [Google Scholar] [CrossRef] [PubMed]
- Kaymaz, S.V.; Ergenç, A.F.; Aytekin, A.; Lucas, S.J.; Elitas, M. A low-cost, portable, and practical LAMP device for point-of-diagnosis in the field. Biotechnol. Bioeng. 2021, 119, 994–1003. [Google Scholar] [CrossRef]
- Yang, J.; Kidd, M.; Nordquist, A.R.; Smith, S.D.; Hurth, C.; Modlin, I.M.; Zenhausern, F. A Sensitive, Portable Microfluidic Device for SARS-CoV-2 Detection from Self-Collected Saliva. Infect. Dis. Rep. 2021, 13, 1061–1077. [Google Scholar] [CrossRef]
- Hu, S.; Jie, Y.; Jin, K.; Zhang, Y.; Guo, T.; Huang, Q.; Mei, Q.; Ma, F.; Ma, H. All-in-One Digital Microfluidics System for Molecular Diagnosis with Loop-Mediated Isothermal Amplification. Biosensors 2022, 12, 324. [Google Scholar] [CrossRef] [PubMed]
- Staples, J.; Dourou, A.-M.; Liampa, I.; Arhondakis, S.; Prakash, R. A Miniaturized Isothermal Rapid DNA Biosensor for SARS-CoV-2 in Clinical and Environmental Samples. In Proceedings of the IEEE Engineering in Medicine and Biology Society Micro and Nanotechnology in Medicine Conference, Kapolei, HI, USA, 5–9 December 2022. [Google Scholar]
- Tharmakulasingam, M.; Chaudhry, N.S.; Branavan, M.; Balachandran, W.; Poirier, A.C.; Rohaim, M.A.; Munir, M.; La Ragione, R.M.; Fernando, A. An Artificial Intelligence-Assisted Portable Low-Cost Device for the Rapid Detection of SARS-CoV-2. Electronics 2021, 10, 2065. [Google Scholar] [CrossRef]
- Malic, L.; Brassard, D.; Da Fonte, D.; Nassif, C.; Mounier, M.; Ponton, A.; Geissler, M.; Shiu, M.; Morton, K.J.; Veres, T. Automated sample-to-answer centrifugal microfluidic system for rapid molecular diagnostics of SARS-CoV-2. Lab A Chip 2022, 22, 3157–3171. [Google Scholar] [CrossRef]
- Prakash, R.; Pabbaraju, K.; Wong, S.; Wong, A.; Tellier, R.; Kaler, K.V.I.S. Multiplex, Quantitative, Reverse Transcription PCR Detection of Influenza Viruses Using Droplet Microfluidic Technology. Micromachines 2014, 6, 63–79. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Z.; Nguyen, N.-T.; Huang, X. Thermocapillary actuation of a water droplet encapsulated in an oil plug. J. Micromech. Microeng. 2007, 17, 1843–1852. [Google Scholar] [CrossRef]
- Karbalaei, A.; Kumar, R.; Cho, H.J. Thermocapillarity in Microfluidics—A Review. Micromachines 2016, 7, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinz, A.; Xing, L.; Doukhanine, E.; Hug, L.A.; Kassen, R.; Ormeci, B.; Kibbee, R.J.; Wong, A.; MacFadden, D.; Nott, C. SARS-CoV-2 detection from the built environment and wastewater and its use for hospital surveillance. Facets 2022, 7, 82–97. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- A Nextflow Pipeline for Running the ARTIC Network’s Field Bioinformatics Tools, with a Focus on ncov2019. Available online: https://github.com/jts/ncov2019-artic-nf (accessed on 7 July 2023).
- Biomarkers Computational Systems: Accelerating Biomarkers Discovery and Optimizing Primers/Probes Design. Available online: https://www.biocos.gr/bioinformatics-2/ (accessed on 9 July 2023).
- PrimerExplorer V5 Software. Available online: http://primerexplorer.jp/lampv5e/index.html (accessed on 8 July 2023).
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef] [PubMed]
- SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests (accessed on 9 July 2023).





| Strain | Lineage | CT Value | Result |
|---|---|---|---|
| Alpha B.1.1 | B.1.1.7 | 20.85, 20.98, 24.04, 24.37, 25.3, 27.0 | 4/6 |
| Delta B.1.6 | B.1.617.2 | 21.19, 21.49, 21.65, 22.81, 26.89, 27.87 | 6/6 |
| Omicron BA.1 | BA.1.1 | 21.60, 23.00, 25.82 | 3/3 |
| Omicron BA.1 | BA.1 | Seven samples with CT in 18–21 | 5/7 |
| Omicron BA.2 | BA.2 | 18.54, 18.94, 19.45, 19.6, 19.67, 21.84, 22.98, 23.31, 23.97, 24.4, 25.37, 25.51, 26.54, 26.82, 33.77 | 12/15 |
| Omicron BA.2 | BA.2.10.1 | 19.71 | 1/1 |
| Omicron BA.4 | BA.4 | 22.26 | 1/1 |
| Omicron BA.4 | BA.4.1 | 16.04, 17.99, 18.39, 23.00 | 3/4 |
| Omicron BA.4 | BA.4.6 | 25.49 | 1/1 |
| Omicron BA.5 | BA.5.1.1 | 17.31 | 1/1 |
| Omicron BA.5 | BA.5.2 | 17.40 | 1/1 |
| Omicron BA.5 | BA.5.2 | 21.01 | 1/1 |
| Omicron BE.1 | BE.1.1 | 20.22 | 1/1 |
| Environmental Swab Samples | Mixed Samples, lineage unknown. (Swabbed between Feb. 2021–Feb. 2022) | 29.26, 30.6, 33.06, 34.29, 38.13, 39.2 | 5/6 |
| LAMP | Primer Sequences |
|---|---|
| BIP | ACT TTC TGT TTT GCT TTC CAT GCA GTT GTA AGG TTG CCC TGT T |
| FIP | CCA TTT TTT CAA AGG CTT CAG TAG TTC ATC TAA ATT GTG GGC TCA |
| B3 | ATG GAA GGG AAC TAA ACT CT |
| F3 | GTT TTG CAA CAA CTC AGA CT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staples, J.; Dourou, A.-M.; Liampa, I.; Sjaarda, C.; Moslinger, E.; Wong, H.; Sheth, P.M.; Arhondakis, S.; Prakash, R. A Miniaturized System for Rapid, Isothermal Detection of SARS-CoV-2 in Human and Environmental Samples. Biomedicines 2023, 11, 2038. https://doi.org/10.3390/biomedicines11072038
Staples J, Dourou A-M, Liampa I, Sjaarda C, Moslinger E, Wong H, Sheth PM, Arhondakis S, Prakash R. A Miniaturized System for Rapid, Isothermal Detection of SARS-CoV-2 in Human and Environmental Samples. Biomedicines. 2023; 11(7):2038. https://doi.org/10.3390/biomedicines11072038
Chicago/Turabian StyleStaples, Jake, Athanasia-Maria Dourou, Irene Liampa, Calvin Sjaarda, Emily Moslinger, Henry Wong, Prameet M. Sheth, Stilianos Arhondakis, and Ravi Prakash. 2023. "A Miniaturized System for Rapid, Isothermal Detection of SARS-CoV-2 in Human and Environmental Samples" Biomedicines 11, no. 7: 2038. https://doi.org/10.3390/biomedicines11072038
APA StyleStaples, J., Dourou, A. -M., Liampa, I., Sjaarda, C., Moslinger, E., Wong, H., Sheth, P. M., Arhondakis, S., & Prakash, R. (2023). A Miniaturized System for Rapid, Isothermal Detection of SARS-CoV-2 in Human and Environmental Samples. Biomedicines, 11(7), 2038. https://doi.org/10.3390/biomedicines11072038