Single-Cell Receptor Quantification of an In Vitro Coculture Angiogenesis Model Reveals VEGFR, NRP1, Tie2, and PDGFR Regulation and Endothelial Heterogeneity
Abstract
:1. Introduction
2. Materials and Methods
3. Results
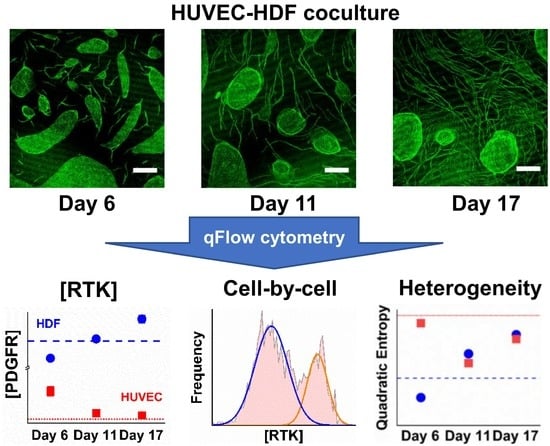
3.1. HUVEC–HDF Cocultures Formed Tubule Networks in a Two-Dimensional Environment
3.2. Co-Culturing Induced a Rapid Increase of VEGFR1 on HDFs within 24 Hours
3.3. Cocultured HUVECs Showed Higher VEGFR2 Concentration than Monoculture throughout Tubule Development
3.4. Tie2 Concentrations on Cocultured HUVECs are Similar to Monocultures
3.5. NRP1 Concentration Decreased on Cocultured Cells within 11 Days
3.6. PDGFRα and PDGFRβ Showed a Steady Increase on Cocultured HDFs from Day 6
3.7. PDGFRβ Observed on Cocultured HUVECs but Not PDGFRα
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
| HUVEC | Human umbilical vein endothelial cell |
| HDF | Human dermal fibroblasts |
| VEGF | Vascular endothelial growth factor |
| Ang | Angiopoietin |
| PDGF | Platelet-derived growth factor |
| FGF | Fibroblast growth factor |
| NRP | Neuropilin |
| PFA | Paraformaldehyde |
| BSA | Bovine serum albumin |
| FBS | Fetal bovine serum |
| DMEM | Dulbecco’s Modified Eagle Media |
| EGM | Endothelial Growth Media |
| EDTA | Ethylenediaminetetraacetic acid |
| HBSS | Hank’s Balanced Salt Solution |
| PE | Phycoerythrin |
| FITC | Fluorescein isothiocyanate |
| DAPI | 4′,6-diamidino-2-phenylindole |
| QE | Quadratic entropy |
| ECM | Extracellular matrix |
| GBM | Glioblastoma |
| Ig | Immunoglobulin |
References
- Folkman, J.; D’Amore, P.A. Blood Vessel Formation: What Is Its Molecular Basis? Cell 1996, 87, 1153–1155. [Google Scholar] [CrossRef] [Green Version]
- Folkman, J. Tumor angiogenesis: Therapeutic implications. N. Engl. J. Med. 1971, 285, 1182–1186. [Google Scholar] [PubMed]
- Imoukhuede, P.I.; Dokun, A.O.; Annex, B.H.; Popel, A.S. Endothelial cell-by-cell profiling reveals the temporal dynamics of VEGFR1 and VEGFR2 membrane localization after murine hindlimb ischemia. Am. J. Physiol. Hear. Circ. Physiol. 2013, 304, H1085–H1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imoukhuede, P.I.; Popel, A.S. Quantitative fluorescent profiling of VEGFRs reveals tumor cell and endothelial cell heterogeneity in breast cancer xenografts. Cancer Med. 2014, 3, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Imoukhuede, P.I.; Popel, A.S. Quantification and cell-to-cell variation of vascular endothelial growth factor receptors. Exp. Cell Res. 2011, 317, 955–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imoukhuede, P.I.; Popel, A.S. Expression of VEGF receptors on endothelial cells in mouse skeletal muscle. PLoS ONE 2012, 7, e44791. [Google Scholar] [CrossRef]
- Chen, S.; Le, T.; Harley, B.A.C.; Imoukhuede, P.I. Characterizing Glioblastoma Heterogeneity via Single-Cell Receptor Quantification. Front. Bioeng. Biotechnol. 2018, 6, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Guo, X.; Imarenezor, O.; Imoukhuede, P.I. Quantification of VEGFRs, NRP1, and PDGFRs on Endothelial Cells and Fibroblasts Reveals Serum, Intra-Family Ligand, and Cross-Family Ligand Regulation. Cell. Mol. Bioeng. 2015, 8, 383–403. [Google Scholar] [CrossRef]
- Weddell, J.C.; Imoukhuede, P.I. Quantitative characterization of cellular membrane-receptor heterogeneity through statistical and computational modeling. PLoS ONE 2014, 9, e97271. [Google Scholar] [CrossRef]
- Finley, S.D.; Engel-Stefanini, M.O.; Imoukhuede, P.I.; Popel, A.S. Pharmacokinetics and pharmacodynamics of VEGF-neutralizing antibodies. BMC Syst. Biol. 2011, 5, 193. [Google Scholar] [CrossRef]
- Gabhann, F.M.; Popel, A.S. Systems Biology of Vascular Endothelial Growth Factors. Microcirculation 2008, 15, 715–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finley, S.D.; Chu, L.-H.; Popel, A.S. Computational systems biology approaches to anti-angiogenic cancer therapeutics. Drug Discov. Today 2014, 20, 187–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qutub, A.A.; Mac Gabhann, F.; Karagiannis, E.D.; Vempati, P.; Popel, A.S. Multiscale models of angiogenesis. IEEE Eng. Med. Biol. Mag. 2009, 28, 14–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mac Gabhann, F.; Qutub, A.A.; Annex, B.H.; Popel, A.S. Systems biology of pro-angiogenic therapies targeting the VEGF system. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Vempati, P.; Popel, A.S.; Mac Gabhann, F. Formation of VEGF isoform-specific spatial distributions governing angiogenesis: Computational analysis. BMC Syst. Biol. 2011, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Vempati, P.; Mac Gabhann, F.; Popel, A.S. Quantifying the proteolytic release of extracellular matrix-sequestered VEGF with a computational model. PLoS ONE 2010, 5, e11860. [Google Scholar] [CrossRef] [PubMed]
- Mac Gabhann, F.; Ji, J.W.; Popel, A.S. VEGF gradients, receptor activation, and sprout guidance in resting and exercising skeletal muscle. J. Appl. Physiol. 2007, 102, 722–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Qutub, A.A.; Vempati, P.; Mac Gabhann, F.; Popel, A.S. Module-based multiscale simulation of angiogenesis in skeletal muscle. Theor. Biol. Med. Model. 2011, 8, 6. [Google Scholar] [CrossRef]
- Qutub, A.A.; Popel, A.S. Elongation, proliferation & migration differentiate endothelial cell phenotypes and determine capillary sprouting. BMC Syst. Biol. 2009, 3, 13. [Google Scholar]
- Ji, J.W.; Mac Gabhann, F.; Popel, A.S. Skeletal muscle VEGF gradients in peripheral arterial disease: Simulations of rest and exercise. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H3740–H3749. [Google Scholar] [CrossRef]
- Weickhardt, A.J.; Williams, D.S.; Lee, C.K.; Chionh, F.; Simes, J.; Murone, C.; Wilson, K.; Parry, M.M.; Asadi, K.; Scott, A.M.; et al. Vascular endothelial growth factor D expression is a potential biomarker of bevacizumab benefit in colorectal cancer. Br. J. Cancer 2015, 113, 37–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirit, M.; Haugh, J.M. Data-driven modelling of receptor tyrosine kinase signalling networks quantifies receptor-specific potencies of PI3K- and Ras-dependent ERK activation. Biochem. J. 2012, 441, 77–85. [Google Scholar] [CrossRef]
- Park, C.S.; Schneider, I.C.; Haugh, J.M. Kinetic analysis of platelet-derived growth factor receptor/phosphoinositide 3-kinase/Akt signaling in fibroblasts. J. Biol. Chem. 2003, 278, 37064–37072. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ansari, A.; Sterrett, W.; Hurley, K.; Kemball, J.; Weddell, J.C.; Imoukhuede, P.I. Current State-of-the-Art and Future Directions in Systems Biology. Prog. Commun. Sci. 2014, 1, 12–26. [Google Scholar]
- Nowak-Sliwinska, P.; Alitalo, K.; Allen, E.; Anisimov, A.; Aplin, A.C.; Auerbach, R.; Augustin, H.G.; Bates, D.O.; van Beijnum, J.R.; Bender, R.H.F.; et al. Consensus guidelines for the use and interpretation of angiogenesis assays. Angiogenesis 2018, 21, 425–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auerbach, R. Angiogenesis Assays: A Critical Overview. Clin. Chem. 2003, 49, 32–40. [Google Scholar] [CrossRef] [Green Version]
- Donovan, D.; Brown, N.J.; Bishop, E.T.; Lewis, C.E. Comparison of three in vitro human ‘angiogenesis’ assays with capillaries formed in vivo. Angiogenesis 2001, 4, 113–121. [Google Scholar] [CrossRef]
- Bishop, E.T.; Bell, G.T.; Bloor, S.; Broom, I.J.; Hendry, N.F.; Wheatley, D.N. An in vitro model of angiogenesis: Basic features. Angiogenesis 1999, 3, 335–344. [Google Scholar] [CrossRef]
- Lin, S.-L.; Kisseleva, T.; Brenner, D.A.; Duffield, J.S. Pericytes and Perivascular Fibroblasts Are the Primary Source of Collagen-Producing Cells in Obstructive Fibrosis of the Kidney. Am. J. Pathol. 2008, 173, 1617–1627. [Google Scholar] [CrossRef] [Green Version]
- Ross, R.; Raines, E.W.; Bowen-Pope, D.F. The biology of platelet-derived growth factor. Cell 1986, 46, 155–169. [Google Scholar] [CrossRef]
- Wolfe, A.; O’Clair, B.; Groppi, V.E.; McEwen, D.P. Pharmacologic Characterization of a Kinetic In Vitro Human Co-Culture Angiogenesis Model Using Clinically Relevant Compounds. J. Biomol. Screen. 2013, 18, 1234–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Weddell, J.C.; Gupta, P.; Conard, G.; Parkin, J.; Imoukhuede, P.I. qFlow cytometry-based receptoromic screening: A high-throughput quantification approach informing biomarker selection and nanosensor development. In Methods in Molecular Biology; Hurst Petrosko, S., Day, E., Eds.; Springer: New York, NY, USA, 2017; pp. 117–138. [Google Scholar]
- Evensen, L.; Micklem, D.R.; Blois, A.; Berge, S.V.; Aarsæther, N.; Littlewood-Evans, A.; Wood, J.; Lorens, J.B. Mural Cell Associated VEGF Is Required for Organotypic Vessel Formation. PLoS ONE 2009, 4, e5798. [Google Scholar] [CrossRef] [PubMed]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaengel, K.; Genové, G.; Armulik, A.; Betsholtz, C. Endothelial-mural cell signaling in vascular development and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, R.F. The aortic ring model of angiogenesis: A quarter century of search and discovery. J. Cell. Mol. Med. 2009, 13, 4113–4136. [Google Scholar] [CrossRef]
- Willett, C.G.; Boucher, Y.; di Tomaso, E.; Duda, D.G.; Munn, L.L.; Tong, R.T.; Chung, D.C.; Sahani, D.V.; Kalva, S.P.; Kozin, S.V.; et al. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat. Med. 2004, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Friis, T.; Kjaer Sorensen, B.; Engel, A.-M.; Rygaard, J.; Houen, G. A quantitative ELISA-based co-culture angiogenesis and cell proliferation assay. APMIS 2003, 111, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Zudaire, E.; Gambardella, L.; Kurcz, C.; Vermeren, S. A computational tool for quantitative analysis of vascular networks. PLoS ONE 2011, 6, e27385. [Google Scholar] [CrossRef]
- Bergers, G.; Song, S.; Meyer-Morse, N.; Bergsland, E.; Hanahan, D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J. Clin. Investig. 2003, 111, 1287–1295. [Google Scholar] [CrossRef] [Green Version]
- Erber, R.; Thurnher, A.; Katsen, A.D.; Groth, G.; Kerger, H.; Hammes, H.-P.; Menger, M.D.; Ullrich, A.; Vajkoczy, P. Combined inhibition of VEGF and PDGF signaling enforces tumor vessel regression by interfering with pericyte-mediated endothelial cell survival mechanisms. FASEB J. 2004, 18, 338–340. [Google Scholar] [CrossRef]
- Casanovas, O.; Hicklin, D.J.; Bergers, G.; Hanahan, D. Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer Cell 2005, 8, 299–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyer, S.; Bishop, J.; Abrams, B.; Maino, V.; Ward, A.; Christian, T.; Davis, K. QuantiBRITE: A new standard for PE flourescence quantitation. In White Paper; Becton Dickinson Immunocytometry Systems: San Jose, CA, USA, 1997. [Google Scholar]
- Kulwinder, K.P.; Edward, T.J.; Sujata, B.I. Performance evaluation of quantiBRITE phycoerythrin beads. Cytometry 2001, 45, 250–258. [Google Scholar]
- Caré, B.R.; Soula, H.A.; Heffetz, D.; Yehiel, Z.; Flörke, R.; Schnaith, K.; Passlack, W.; Wichert, M.; Kuehn, L.; Fabry, M.; et al. Impact of receptor clustering on ligand binding. BMC Syst. Biol. 2011, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Mahama, P.A.; Linderman, J.J. A Monte Carlo study of the dynamics of G-protein activation. Biophys. J. 1994, 67, 1345–1357. [Google Scholar] [CrossRef] [Green Version]
- Shibuya, M. Vascular endothelial growth factor receptor-1 (VEGFR-1/Flt-1): A dual regulator for angiogenesis. Angiogenesis 2006, 9, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, M.O.; Wu, F.T.H.; Mac Gabhann, F.; Popel, A.S. Increase of plasma VEGF after intravenous administration of bevacizumab is predicted by a pharmacokinetic model. Cancer Res. 2010, 70, 9886–9894. [Google Scholar] [CrossRef]
- Weddell, J.C.; Chen, S.; Imoukhuede, P.I. VEGFR1 promotes cell migration and proliferation through PLCγ and PI3K pathways. NPJ Syst. Biol. Appl. 2018, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.R. Diversity and dissimilarity coefficients: A unified approach. Theor. Popul. Biol. 1982, 21, 24–43. [Google Scholar] [CrossRef]
- Pavoine, S.; Dolédec, S. The apportionment of quadratic entropy: A useful alternative for partitioning diversity in ecological data. Environ. Ecol. Stat. 2005, 12, 125–138. [Google Scholar] [CrossRef]
- Botta-Dukát, Z. Rao’s quadratic entropy as a measure of functional diversity based on multiple traits. J. Veg. Sci. 2005, 16, 533–540. [Google Scholar] [CrossRef]
- Olsson, A.-K.; Dimberg, A.; Kreuger, J.; Claesson-Welsh, L. VEGF receptor signalling—In control of vascular function. Nat. Rev. Mol. Cell Biol. 2006, 7, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Koch, S.; Claesson-Welsh, L. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb. Perspect. Med. 2012, 2, a006502. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, S.; Sako, K.; Minami, T.; Noda, K.; Kim, H.Z.; Kodama, T.; Shibuya, M.; Takakura, N.; Koh, G.Y.; Mochizuki, N. Differential function of Tie2 at cell–cell contacts and cell–substratum contacts regulated by angiopoietin-1. Nat. Cell Biol. 2008, 10, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Hellström, M.; Kalén, M.; Lindahl, P.; Abramsson, A.; Betsholtz, C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 1999, 126, 3047–3055. [Google Scholar] [PubMed]
- Zhang, J.; Cao, R.; Zhang, Y.; Jia, T.; Cao, Y.; Wahlberg, E. Differential roles of PDGFR-alpha and PDGFR-beta in angiogenesis and vessel stability. FASEB J. 2009, 23, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Raines, E.W.; Bowen-Pope, D.F.; Ross, R. Platelet-Derived Growth Factor. In Peptide Growth Factors and Their Receptors I; Springer: New York, NY, USA, 1991; pp. 173–262. [Google Scholar]
- Lindner, V.; Reidy, M.A. Platelet-derived growth factor ligand and receptor expression by large vessel endothelium in vivo. Am. J. Pathol. 1995, 146, 1488–1497. [Google Scholar] [PubMed]
- Gerhardt, H.; Golding, M.; Fruttiger, M.; Ruhrberg, C.; Lundkvist, A.; Abramsson, A.; Jeltsch, M.; Mitchell, C.; Alitalo, K.; Shima, D.; et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol. 2003, 161, 1163–1177. [Google Scholar] [CrossRef]
- Battegay, E.J.; Rupp, J.; Iruela-Arispe, L.; Sage, E.H.; Pech, M. PDGF-BB modulates endothelial proliferation and angiogenesis in vitro via PDGF beta-receptors. J. Cell Biol. 1994, 125, 917–928. [Google Scholar] [CrossRef]
- Smits, A.; Hermansson, M.; Nister, M.; Karnushina, I.; Heldin, C.-H.; Westermark, B.; Funa, K. Rat Brain Capillary Endothelial Cells Express Functional PDGF B-Type Receptors. Growth Factors 1989, 2, 1–8. [Google Scholar] [CrossRef]
- Stratman, A.N.; Malotte, K.M.; Mahan, R.D.; Davis, M.J.; Davis, G.E. Pericyte recruitment during vasculogenic tube assembly stimulates endothelial basement membrane matrix formation. Blood 2009, 114, 5091–5101. [Google Scholar] [CrossRef] [Green Version]
- Sieveking, D.P.; Buckle, A.; Celermajer, D.S.; Ng, M.K.C. Strikingly different angiogenic properties of endothelial progenitor cell subpopulations: Insights from a novel human angiogenesis assay. J. Am. Coll. Cardiol. 2008, 51, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Bryan, B.A.; D’Amore, P.A. Pericyte isolation and use in endothelial/pericyte coculture models. Methods Enzymol. 2008, 443, 315–331. [Google Scholar] [PubMed]
- Armulik, A.; Genové, G.; Betsholtz, C. Pericytes: Developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 2011, 21, 193–215. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Shibuya, M. The 230 kDa mature form of KDR/Flk-1 (VEGF receptor-2) activates the PLC-γ pathway and partially induces mitotic signals in NIH3T3 fibroblasts. Oncogene 1997, 14, 2079–2089. [Google Scholar] [CrossRef] [PubMed]
- Berthaut, A.; Mirshahi, P.; Benabbou, N.; Azzazene, D.; Bordu, C.; Therwath, A.; Legeais, J.; Mirshahi, M. Vascular endothelial growth factor receptor-1 (VEGFR-1) expression in human corneal fibroblast decreased with age. Mol. Vis. 2009, 15, 1997–2007. [Google Scholar] [PubMed]
- Itoh, H.; Nasu, K.; Matsumoto, H.; Kawano, Y.; Yoshimatsu, J.; Narahara, H. Hypoxia regulates vascular endothelial growth factor and soluble fms-like tyrosine kinase-1 secretion by human oviductal epithelial cells and stromal fibroblasts. Fertil. Steril. 2006, 85, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M.; Claesson-Welsh, L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Exp. Cell Res. 2006, 312, 549–560. [Google Scholar] [CrossRef]
- Siemerink, M.J.; Klaassen, I.; Van Noorden, C.J.F.; Schlingemann, R.O. Endothelial tip cells in ocular angiogenesis: Potential target for anti-angiogenesis therapy. J. Histochem. Cytochem. 2013, 61, 101–115. [Google Scholar] [CrossRef]
- Adams, R.H.; Alitalo, K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol. 2007, 8, 464–478. [Google Scholar] [CrossRef]
- Eklund, L.; Olsen, B.R. Tie receptors and their angiopoietin ligands are context-dependent regulators of vascular remodeling. Exp. Cell Res. 2006, 312, 630–641. [Google Scholar] [CrossRef]
- Saharinen, P.; Eklund, L.; Miettinen, J.; Wirkkala, R.; Anisimov, A.; Winderlich, M.; Nottebaum, A.; Vestweber, D.; Deutsch, U.; Koh, G.Y.; et al. Angiopoietins assemble distinct Tie2 signalling complexes in endothelial cell-cell and cell-matrix contacts. Nat. Cell Biol. 2008, 10, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Goettsch, W.; Gryczka, C.; Korff, T.; Ernst, E.; Goettsch, C.; Seebach, J.; Schnittler, H.-J.; Augustin, H.G.; Morawietz, H. Flow-dependent regulation of angiopoietin-2. J. Cell. Physiol. 2008, 214, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Obi, S.; Masuda, H.; Shizuno, T.; Sato, A.; Yamamoto, K.; Ando, J.; Abe, Y.; Asahara, T. Fluid shear stress induces differentiation of circulating phenotype endothelial progenitor cells. Am. J. Physiol. Physiol. 2012, 303, C595–C606. [Google Scholar] [CrossRef] [PubMed]
- Kelly-Goss, M.R.; Ning, B.; Bruce, A.C.; Tavakol, D.N.; Yi, D.; Hu, S.; Yates, P.A.; Peirce, S.M. Dynamic, heterogeneous endothelial Tie2 expression and capillary blood flow during microvascular remodeling. Sci. Rep. 2017, 7, 9049. [Google Scholar] [CrossRef] [PubMed]
- Biel, N.M.; Siemann, D.W. Targeting the Angiopoietin-2/Tie-2 axis in conjunction with VEGF signal interference. Cancer Lett. 2016, 380, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Bhat, A.; Woodnutt, G.; Lappe, R. Targeting the ANGPT–TIE2 pathway in malignancy. Nat. Rev. Cancer 2010, 10, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Hirschi, K.K.; Rohovsky, S.A.; D’Amore, P.A. PDGF, TGF-beta, and Heterotypic Cell-Cell Interactions Mediate Endothelial Cell-induced Recruitment of 10T1/2 Cells and Their Differentiation to a Smooth Muscle Fate. J. Cell Biol. 1998, 141, 805–814. [Google Scholar] [CrossRef]
- Cobbs, C.; Khan, S.; Matlaf, L.; McAllister, S.; Zider, A.; Yount, G.; Rahlin, K.; Harkins, L.; Bezrookove, V.; Singer, E.; et al. HCMV glycoprotein B is expressed in primary glioblastomas and enhances growth and invasiveness via PDGFR-alpha activation. Oncotarget 2014, 5, 1091–1100. [Google Scholar] [CrossRef]
- Bhardwaj, B.; Klassen, J.; Cossette, N.; Sterns, E.; Tuck, A.; Deeley, R.; Sengupta, S.; Elliott, B. Localization of platelet-derived growth factor beta receptor expression in the periepithelial stroma of human breast carcinoma. Clin. Cancer Res. 1996, 2, 773–782. [Google Scholar]
- Krupinski, J.; Issa, R.; Bujny, T.; Slevin, M.; Kumar, P.; Kumar, S.; Kaluza, J. A Putative Role for Platelet-Derived Growth Factor in Angiogenesis and Neuroprotection After Ischemic Stroke in Humans. Stroke 1997, 28, 564–573. [Google Scholar] [CrossRef]
- Aird, W.C. Molecular heterogeneity of tumor endothelium. Cell Tissue Res. 2009, 335, 271–281. [Google Scholar] [CrossRef]
- Aird, W.C. Spatial and temporal dynamics of the endothelium. J. Thromb. Haemost. 2005, 3, 1392–1406. [Google Scholar] [CrossRef] [PubMed]
- Passarelli, M.K.; Ewing, A.G. Single-cell imaging mass spectrometry. Curr. Opin. Chem. Biol. 2013, 17, 854–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, S.; Zeisel, A.; Joost, S.; La Manno, G.; Zajac, P.; Kasper, M.; Lönnerberg, P.; Linnarsson, S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 2014, 11, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.R.; Ribas, A.; Mischel, P.S. Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov. 2016, 15, 204–216. [Google Scholar] [CrossRef]
- Shi, Q.; Qin, L.; Wei, W.; Geng, F.; Fan, R.; Shin, Y.S.; Guo, D.; Hood, L.; Mischel, P.S.; Heath, J.R. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl. Acad. Sci. USA 2012, 109, 419–424. [Google Scholar] [CrossRef]
- Staton, C.A.; Stribbling, S.M.; Tazzyman, S.; Hughes, R.; Brown, N.J.; Lewis, C.E. Current methods for assaying angiogenesis in vitro and in vivo. Int. J. Exp. Pathol. 2004, 85, 233–248. [Google Scholar] [CrossRef]
- Staton, C.A.; Reed, M.W.R.; Brown, N.J. A critical analysis of current in vitro and in vivo angiogenesis assays. Int. J. Exp. Pathol. 2009, 90, 195–221. [Google Scholar] [CrossRef]
- Styp-Rekowska, B.; Hlushchuk, R.; Pries, A.R.; Djonov, V. Intussusceptive angiogenesis: Pillars against the blood flow. Acta Physiol. 2011, 202, 213–223. [Google Scholar] [CrossRef]
- Dela Paz, N.G.; Walshe, T.E.; Leach, L.L.; Saint-Geniez, M.; D’Amore, P.A. Role of shear-stress-induced VEGF expression in endothelial cell survival. J. Cell Sci. 2012, 125, 831–843. [Google Scholar] [CrossRef] [Green Version]
- Moya, M.L.; Hsu, Y.-H.; Lee, A.P.; Hughes, C.C.W.; George, S.C. In Vitro Perfused Human Capillary Networks. Tissue Eng. Part C Methods 2013, 19, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Grassot, J.; Gouy, M.; Perrière, G.; Mouchiroud, G. Origin and Molecular Evolution of Receptor Tyrosine Kinases with Immunoglobulin-Like Domains. Mol. Biol. Evol. 2006, 23, 1232–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarabipour, S.; Hristova, K. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 2016, 7, 10262. [Google Scholar] [CrossRef] [PubMed]
- Taeger, J.; Moser, C.; Hellerbrand, C.; Mycielska, M.E.; Glockzin, G.; Schlitt, H.J.; Geissler, E.K.; Stoeltzing, O.; Lang, S.A.; Jemal, A.; et al. Targeting FGFR/PDGFR/VEGFR impairs tumor growth, angiogenesis, and metastasis by effects on tumor cells, endothelial cells, and pericytes in pancreatic cancer. Mol. Cancer Ther. 2011, 10, 2157–2167. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Imoukhuede, P.I. Single-Cell Receptor Quantification of an In Vitro Coculture Angiogenesis Model Reveals VEGFR, NRP1, Tie2, and PDGFR Regulation and Endothelial Heterogeneity. Processes 2019, 7, 356. https://doi.org/10.3390/pr7060356
Chen S, Imoukhuede PI. Single-Cell Receptor Quantification of an In Vitro Coculture Angiogenesis Model Reveals VEGFR, NRP1, Tie2, and PDGFR Regulation and Endothelial Heterogeneity. Processes. 2019; 7(6):356. https://doi.org/10.3390/pr7060356
Chicago/Turabian StyleChen, Si, and P. I. Imoukhuede. 2019. "Single-Cell Receptor Quantification of an In Vitro Coculture Angiogenesis Model Reveals VEGFR, NRP1, Tie2, and PDGFR Regulation and Endothelial Heterogeneity" Processes 7, no. 6: 356. https://doi.org/10.3390/pr7060356
APA StyleChen, S., & Imoukhuede, P. I. (2019). Single-Cell Receptor Quantification of an In Vitro Coculture Angiogenesis Model Reveals VEGFR, NRP1, Tie2, and PDGFR Regulation and Endothelial Heterogeneity. Processes, 7(6), 356. https://doi.org/10.3390/pr7060356