Horseradish Peroxidase-Decorated Artificial Viral Capsid Constructed from β-Annulus Peptide via Interaction between His-Tag and Ni-NTA
Abstract
:1. Introduction
2. Materials and Methods
2.1. General
2.2. Preparetion of His-Tagged HRP
2.3. Preparation of β-Annulus Peptide C-Terminus-Modified with Ni-NTA
2.4. Complexation of His-Tagged HRP with Ni-NTA-Modified β-Annulus Peptide
2.5. Dynamic Light Scattering (DLS) and ζ-Potential
2.6. Transmission Electron Microscopy (TEM)
2.7. HRP Activity
3. Results
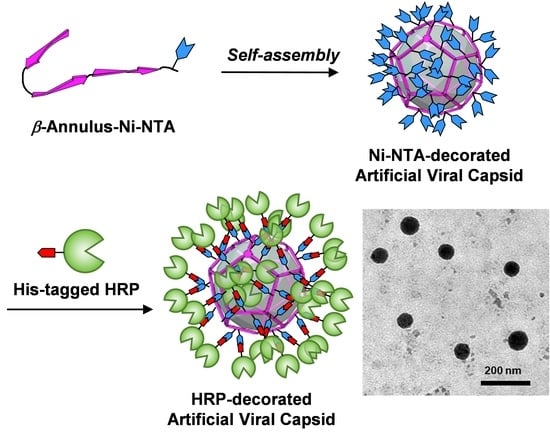
3.1. Construction of an Artificial Viral Capsid Modified with Ni-NTA at the Surface
3.2. Preparation of His-Tagged HRP
3.3. Construction of HRP-Decorated Artificial Viral Capsid
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Harper, D.R. Viruses: Biology, Applications, and Control, Garland Science, 2nd ed.; Taylor & Francis Group: Oxfordshire, UK, 2012. [Google Scholar]
- Mateu, M.G. (Ed.) Structure and Physics of Viruses; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Brodsky, F.M. Diversity of clathrin function: New tricks for an old protein. Annu. Rev. Cell Dev. Biol. 2012, 28, 309–336. [Google Scholar] [CrossRef] [PubMed]
- Yeates, T.O.; Kerfeld, C.A.; Heinhorst, S.; Cannon, G.C.; Shively, J.M. Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat. Rev. Microbiol. 2008, 6, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Giessen, T.W.; Silver, P.A. Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat. Microbiol. 2017, 2, 17029. [Google Scholar] [CrossRef] [PubMed]
- Gabashvili, A.N.; Chmelyuk, N.S.; Efremova, M.V.; Malinovskaya, J.A.; Semkina, A.S.; Abakumov, M.A. Encapsulins—Bacterial protein nanocompartments: Structure, properties, and application. Biomolecules 2020, 10, 966. [Google Scholar] [CrossRef]
- Azuma, Y.; Edwardson, T.G.W.; Hilvert, D. Tailoring lumazine synthase assemblies for bionanotechnology. Chem. Soc. Rev. 2018, 47, 3543–3557. [Google Scholar] [CrossRef]
- Jutz, G.; van Rijn, P.; Miranda, B.S.; Böker, A. Ferritin: A versatile building block for bionanotechnology. Chem. Rev. 2015, 115, 1653–1701. [Google Scholar] [CrossRef]
- Ueno, T.; Tabe, H.; Tanaka, Y. Artificial metalloenzymes constructed from hierarchically-assembled proteins. Chem. Asian J. 2013, 8, 1646–1660. [Google Scholar] [CrossRef] [PubMed]
- Khudyakov, Y.; Pumpens, P. (Eds.) Viral Nanotechnology; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Wen, A.M.; Steinmetz, N.F. Design of virus-based nanomaterials for medicine, biotechnology, and energy. Chem. Soc. Rev. 2016, 45, 4074–4126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Rijn, P.; Schirhagl, R. Viruses, artificial viruses and virus-based structures for biomedical applications. Adv. Healthcare Mater. 2016, 5, 1386–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhaskar, S.; Lim, S. Engineering protein nanocages as carriers for biomedical applications. NPG Asia Mater. 2017, 9, e371. [Google Scholar] [CrossRef]
- Ren, H.; Zhu, S.; Zheng, G. Nanoreactor design based on self-assembling protein nanocages. Int. J. Mol. Sci. 2019, 20, 592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maity, B.; Fujita, K.; Ueno, T. Use of the confined spaces of apo-ferritin and virus capsids as nanoreactors for catalytic reactions. Curr. Opin. Chem. Biol. 2015, 25, 88–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuurua, K. Rational design of self-assembled proteins and peptides for nano- and micro-sized architectures. RSC Adv. 2014, 4, 2942–2953. [Google Scholar] [CrossRef]
- De Santis, E.; Ryadnov, M.G. Peptide self-assembly for nanomaterials: The old new kid on the block. Chem. Soc. Rev. 2015, 44, 8288–8300. [Google Scholar] [CrossRef]
- Luo, Q.; Hou, C.; Bai, Y.; Wang, R.; Liu, J. Protein assembly: Versatile approaches to construct highly ordered nanostructures. Chem. Rev. 2016, 116, 13571–13632. [Google Scholar] [CrossRef]
- Matsuura, K. Synthetic approaches to construct viral capsid-like spherical nanomaterials. Chem. Commun. 2018, 54, 8944–8959. [Google Scholar] [CrossRef]
- Kuan, S.L.; Bergamini, F.R.G.; Weil, T. Functional protein nanostructures: A chemical toolbox. Chem. Soc. Rev. 2018, 47, 9069–9105. [Google Scholar] [CrossRef] [Green Version]
- Lou, S.; Wang, X.; Yu, Z.; Shi, L. Peptide tectonics: Encoded structural complementarity dictates programmable self-assembly. Adv. Sci. 2019, 6, 1802043. [Google Scholar] [CrossRef] [Green Version]
- Hamley, I.W. Protein assemblies: Nature-inspired and designed nanostructures. Biomacromolecules 2019, 20, 1829–1848. [Google Scholar] [CrossRef] [Green Version]
- Boerakker, M.J.; Hannink, J.M.; Bomans, P.H.H.; Frederik, P.M.; Nolte, R.J.M.; Meijer, E.M.; Sommerdijk, N.A.J.M. Giant amphiphiles by cofactor reconstitution. Angew. Chem. Int. Ed. 2002, 41, 4239–4241. [Google Scholar] [CrossRef]
- Reynhout, I.C.; Cornelissen, J.J.L.M.; Nolte, R.J.M. Self-assembled architectures from biohybrid triblock copolymers. J. Am. Chem. Soc. 2007, 129, 2327–2332. [Google Scholar] [CrossRef] [PubMed]
- Delaittre, G.; Reynhout, I.C.; Cornelissen, J.J.L.M.; Nolte, R.J.M. Cascade reactions in an all-enzyme nanoreactor. Chem. Eur. J. 2009, 15, 12600–12603. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, S.; Oohora, K.; Uchihashi, T.; Hayashi, T. Thermoresponsive micellar assembly constructed from a hexameric hemoprotein modified with poly(N-isopropylacrylamide) toward an artificial light-harvesting system. J. Am. Chem. Soc. 2020, 142, 1822–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fletcher, J.M.; Harniman, R.L.; Barnes, F.R.H.; Boyle, A.L.; Collins, A.; Mantell, J.; Sharp, T.H.; Antognozzi, M.; Booth, P.J.; Linden, N.; et al. Self-assembling cages from coiled-coil peptide modules. Science 2013, 340, 595–599. [Google Scholar] [CrossRef]
- Ross, J.F.; Bridges, A.; Fletcher, J.M.; Shoemark, D.; Alibhai, D.; Bray, H.E.V.; Beesley, J.L.; Dawson, W.M.; Hodgson, L.R.; Mantell, J.; et al. Decorating self-assembled peptide cages with proteins. ACS Nano 2017, 11, 7901–7914. [Google Scholar] [CrossRef]
- Kawakami, N.; Kondo, H.; Matsuzawa, Y.; Hayasaka, K.; Nasu, E.; Sasahara, K.; Arai, R.; Miyamoto, K. Design of hollow protein nanoparticles with modifiable interior and exterior surfaces. Angew. Chem. Int. Ed. 2018, 57, 12400–12404. [Google Scholar] [CrossRef]
- Cristie-David, A.S.; Chen, J.; Nowak, D.B.; Bondy, A.L.; Sun, K.; Park, S.I.; Holl, M.M.B.; Su, M.; Marsh, E.N.G. Coiled-coil-mediated assembly of an icosahedral protein cage with extremely high thermal and chemical stability. J. Am. Chem. Soc. 2019, 141, 9207–9216. [Google Scholar] [CrossRef]
- De Santis, E.; Alkassem, H.; Lamarre, B.; Faruqui, N.; Bella, A.; Noble, J.E.; Micale, N.; Ray, S.; Burns, J.R.; Yon, A.R.; et al. Antimicrobial peptide capsids of de novo design. Nat. Commun. 2017, 8, 2263. [Google Scholar] [CrossRef] [Green Version]
- Kepiro, I.E.; Marzuoli, I.; Hammond, K.; Ba, X.; Lewis, H.; Shaw, M.; Gunnoo, S.B.; De Santis, E.; Łapińska, U.; Pagliara, S.; et al. Engineering chirally blind protein pseudocapsids into antibacterial persisters. ACS Nano 2020, 14, 1609–1622. [Google Scholar] [CrossRef]
- Matsuura, K.; Watanabe, K.; Sakurai, K.; Matsuzaki, T.; Kimizuka, N. Self-assembled synthetic viral capsids from a 24-mer viral peptide fragment. Angew. Chem. Int. Ed. 2010, 49, 9662–9665. [Google Scholar] [CrossRef]
- Matsuura, K.; Watanabe, K.; Matsushita, Y.; Kimizuka, N. Guest-binding behavior of peptide nanocapsules self-assembled from viral peptide fragments. Polymer J. 2013, 45, 529–534. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Inaba, H.; Matsuura, K. Construction of artificial viral capsids encapsulating short DNAs via disulfide bonds and controlled release of DNAs by reduction. Chem. Lett. 2019, 48, 544–546. [Google Scholar] [CrossRef]
- Fujita, S.; Matsuura, K. Inclusion of zinc oxide nanoparticles into virus-like peptide nanocapsules self-assembled from viral β-annulus peptide. Nanomaterials 2014, 4, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Fujita, S.; Matsuura, K. Encapsulation of CdTe quantum dots into synthetic viral capsids. Chem. Lett. 2016, 45, 922–924. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, K.; Nakamura, T.; Watanabe, K.; Noguchi, T.; Minamihata, K.; Kamiya, N.; Kimizuka, N. Self-assembly of Ni-NTA-modified β-annulus peptides into artificial viral capsids and encapsulation of His-tagged proteins. Org. Biomol. Chem. 2016, 14, 7869–7874. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, K.; Ueno, G.; Fujita, S. Self-assembled artificial viral capsid decorated with gold nanoparticles. Polymer J. 2015, 47, 146–151. [Google Scholar] [CrossRef] [Green Version]
- Fujita, S.; Matsuura, K. Self-assembled artificial viral capsids bearing coiled-coils at the surface. Org. Biomol. Chem. 2017, 15, 5070–5077. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Yamada, S.; Nishikawa, S.; Matsuura, K. DNA-modified artificial viral capsids self-assembled from DNA-conjugated β-annulus peptide. J. Pept. Sci. 2017, 23, 636–643. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, K.; Honjo, T. Artificial viral capsid dressed up with human serum albumin. Bioconj. Chem. 2019, 30, 1636–1641. [Google Scholar] [CrossRef]
- Matsuura, K.; Ota, J.; Fujita, S.; Shiomi, Y.; Inaba, H. Construction of ribonuclease-decorated artificial virus-like capsid by peptide self-assembly. J. Org. Chem. 2020, 85, 1668–1673. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, K. Dressing up artificial viral capsids self-assembled from C-terminal-modified β-annulus peptides. Polymer J. 2020, 52, 1035–1041. [Google Scholar] [CrossRef]
- Grigorenko, V.; Chuber, T.; Kapeliuch, Y.; Borchers, T.; Spener, F.; Egorov, A. New approaches for functional expression of recombinant horseradish peroxidase C in Escherichia coli. Biocatal. Biotransfor. 1999, 17, 359–379. [Google Scholar] [CrossRef]
- Nicell, J.A.; Wright, H. A model of peroxidase activity with inhibition by hydrogen peroxide. Enzym. Microb. Technol. 1997, 21, 302–310. [Google Scholar] [CrossRef]
- Dinh, H.; Nakata, E.; Mutsuda-Zapater, K.; Saimura, M.; Kinoshita, M.; Morii, T. Enhanced enzymatic activity exerted by a packed assembly of a single type of enzyme. Chem. Sci. 2020, 11, 9088–9100. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuura, K.; Shiomi, Y.; Mizuta, T.; Inaba, H. Horseradish Peroxidase-Decorated Artificial Viral Capsid Constructed from β-Annulus Peptide via Interaction between His-Tag and Ni-NTA. Processes 2020, 8, 1455. https://doi.org/10.3390/pr8111455
Matsuura K, Shiomi Y, Mizuta T, Inaba H. Horseradish Peroxidase-Decorated Artificial Viral Capsid Constructed from β-Annulus Peptide via Interaction between His-Tag and Ni-NTA. Processes. 2020; 8(11):1455. https://doi.org/10.3390/pr8111455
Chicago/Turabian StyleMatsuura, Kazunori, Yuriko Shiomi, Toshihumi Mizuta, and Hiroshi Inaba. 2020. "Horseradish Peroxidase-Decorated Artificial Viral Capsid Constructed from β-Annulus Peptide via Interaction between His-Tag and Ni-NTA" Processes 8, no. 11: 1455. https://doi.org/10.3390/pr8111455
APA StyleMatsuura, K., Shiomi, Y., Mizuta, T., & Inaba, H. (2020). Horseradish Peroxidase-Decorated Artificial Viral Capsid Constructed from β-Annulus Peptide via Interaction between His-Tag and Ni-NTA. Processes, 8(11), 1455. https://doi.org/10.3390/pr8111455