Identification and Isolation Techniques for Plant Growth Inhibitors in Rice
Abstract
:1. Introduction
2. Literature Sources and Search Methodology
3. Mechanism of Action of PGIs
3.1. Oxidative Stress
3.2. Cell Division and Permeability
3.3. Photosynthesis, Respiration, and Transpiration
3.4. Gene Expression, Protein Biosynthesis, Phytohormone Activities, and Enzyme Functions
3.5. Symbiotic and Pathogenic Microorganisms
4. Major PGIs in Rice
4.1. Phenolic Acids
4.2. Polyphenols
4.3. Terpenoids
4.4. Other PGIs
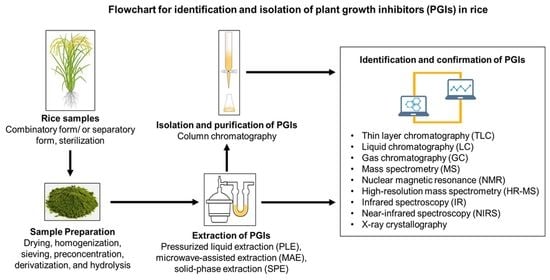
5. Isolation and Identification Techniques for PGIs in Rice
5.1. Preparation of Rice Samples
5.2. Extraction, Separation, Isolation, and Purification Techniques
5.2.1. Extraction and Separation Techniques
| Rice Samples | Compounds | Techniques | Conditions | Tested Plants | Ref |
|---|---|---|---|---|---|
| Rice plants | (S)-Limonene | SPME | Solid phase (SPME): divinylbenzene (DVB)/carboxen (CAR)/polydimethylsiloxane (PDMS) | Bacterial blight (Xanthomonas oryzae pv. oryzae) | [20] |
| Rice husk | Momilactones A, B, E, and 7-Ketostigmasterol | Repeated-column chromatography | Hexan:EtOAc; CHCl3:MeOH Silica gel 60–100 mesh size in a 5 × 60 cm column followed by 200–400 mesh in a 2 × 50 cm column | Barnyardgrass (E. crus-galli), tall goldenrod (S. altissima), and Lettuce (Lactuca sativa) | [99] |
| Rice grains | Phenolics | UAE | MeOH:H2O (80:20, v/v) At 45 °C, in 25 min, cycle 0.4 s−1, ultrasound amplitude 47% | Wild grass | [132] |
| Rice grains | Phenolics | PLE | EtOAc:MeOH (60:40, v/v) Pressurized 200 atm and then heated 190 °C in 10 min with three cycles | Wild grass | [133] |
| Rice grains | Phenolics | MAE | 100% MeOH Solve to sample (10:1) At 185 °C, 20 min, cycle 0.4 s−1, microwave power 1000 W | Wild grass | [134] |
5.2.2. Isolation and Purification Techniques
5.3. Qualitative and Quantitative Analyses
5.3.1. Spectroscopic Methods
5.3.2. Chromatography
| Rice Samples | Compounds | Techniques | Conditions | Ref. | ||
|---|---|---|---|---|---|---|
| LC Techniques | Stationary Phase | Technical Index | Mobile Phase | |||
| Rice hulls | Diterpenoids | TLC-NMR/ESI/MS | Si-gel G 60 F254 plates | CHCl3 | [22] | |
| Rice hulls | Momilactones A, B, E, and 7-Ketostigmasterol | NMR/ESI/HR/MS-HPLC | Waters Spherisorb ODS2 column | 150 mm × 4.6 mm, 10 µm | 0.1% Trifluoroacetic acid (TFA) in 70% ACN | [99] |
| Rice grains | Momilactones A and B | UPLC-ESI-MS/MS | UPLC® BEH C18 column | 1.7 µm, 50 × 2.1 mm | 0.1% TFA and 0.1% TFA in ACN (50:50, v/v) in a gradient program | [26] |
| Rice germplasm | Flavone O-glycosides | HPLC | Zorbax SB-C18 column | 150 mm × 4.6 mm, 5 μm | ACN:AcOH (1%): 2:3 (v/v) | [46] |
| Shoots | Jasmonic acid | UPLC-MS/MS | BEHC18 column | 50 mm × 2.1 mm, 1.7 μm | AcOH in H2O (0.2%): MeOH Flow rate of 300 μL/min | [161] |
| Rice cultivars | 2,4-Dimethoxybenzoic acid, p-coumaric acid, vanillic acid, salicylic acid, and cinnamic acid. | HPLC-UV/Vis | XDB-C18 column | 150 mm × 4.6 mm, 5 μm equipped with 20 mm × 3.9 mm, 5 μm | A: MeOH/ B: HCOOH 0.1% A:B (30:30, v/v) or A: ACN/ B: HCOOH 0.1% A:B (20:80, v/v) | [36] |
| Rice leaves | Phenolics and phytoalexins | LC–MS/MS | Shiseido Capcell PaK C8 column | 150 mm × 4.6 mm, 5 μm | ACN (HCOOH 0.1%):D.I.W. (80:20, v/v) | [162] |
| Rice leaves | Phenylamides | LC-MS/MS-NMR | Acquity UPLC BEH C18 column | 50 mm × 2.1 mm, 1.7 μm | A: HCOOH 0.1% aqueous B: HCOOH 0.1% ACN | [108] |
| Rice bran | Phenolics | HPLC-DAD | Kinetex C18 column | 150 mm × 4.6 mm, 5 μm | A: D.I.W (HCOOH 0.1%). B: ACN (HCOOH 0.1%). gradient 5–70% A/(A + B) | [163] |
| Non-glutinous purple rice | Phenolics | HPLC-ESI-MS/MS | Zorbax Eclipse XDB C18 column | 150 mm × 4.6 mm, 5 µm |
at pH 2.5 B: ACN
B: MeOH | [152] |
| GC Techniques | Conditions | Ref. | ||||
| Rice plants | (S)-Limonene | SPME- GC/MS |
| [20] | ||
| Rice leaves | Oryzalides and oryzalic acids | GC-SIM |
| [157] | ||
| Rice plants | Flavone O-glycoside | GC-FID |
| [158] | ||
| Japanese rice | Momilactones A and B | GC/MS |
| [32] | ||
| Black and purple rice bran | Flavonoids | UAE-GC/MS/FID |
| [160] | ||
6. Current Status, Existing Limitations, and Future Perspectives
6.1. Current Status and Existing Limitations
6.1.1. Extraction and Separation Techniques
6.1.2. Identification, Qualitative and Quantitative Analyses
6.2. Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAO. Food Outlook Biannual Report on Global Food Markets; FAO: Rome, Italy, 2022. [Google Scholar]
- FAO. Rice is life. In Proceedings of the FAO Rice Conference, Rome, Italy, 12–13 February 2004. [Google Scholar]
- Hoed, V.V.; Depaemelaere, G.; Ayala, J.V.; Santiwattana, P.; Verhe, R.; Greyt, W.D. Influence of chemical refining on the major and minor components of rice brain oil. J. Am. Oil Chem. Soc. 2006, 83, 315–321. [Google Scholar] [CrossRef]
- Friedman, M. Rice brans, rice bran oils, and rice hulls: Composition, food and industrial uses, and bioactivities in humans, animals, and cells. J. Agric. Food Chem. 2013, 61, 10626–10641. [Google Scholar] [CrossRef] [PubMed]
- Tadahiro, K.; Chizuko, K.; Nobuki, S.; Mitsuaki, T.; Hiroyasu, A.; Kenichi, F.; Yoshiaki, K.; Yoshio, K.; Norindo, T. Momilactones, growth inhibitors from rice, Oryza sativa L. Tetrahedron Lett. 1973, 14, 3861–3864. [Google Scholar]
- Amb, M.K.; Ahluwalia, A.S. Allelopathy: Potential role to achieve new milestones in rice cultivation. Rice Sci. 2016, 23, 165–183. [Google Scholar] [CrossRef]
- Seung, W.L.; Udo, B.; Thomas, M.G.; Timothy, E.O. Effects of mixtures of phenolic acids on phosphorus uptake by cucumber seedlings. J. Chem. Ecol. 1990, 16, 2559–2567. [Google Scholar]
- Kuwatsuka, S.; Shindo, H. Behavior of phenolic substances in the decaying process of plants: Identification and quantitative determination of phenolic acids in rice straw and its decayed product by gas chromatography. J. Soil Sci. Plant Nutr. 1973, 19, 219–227. [Google Scholar] [CrossRef]
- Park, M.H.; Kim, B.H.; Chung, I.M.; Hwang, S.J. Selective bactericidal potential of rice (Oryza sativa L. var. japonica) hull extract on microcystis strains in comparison with green algae and Zooplankton. Bull. Environ. Contam. Toxicol. 2009, 83, 97–101. [Google Scholar] [CrossRef]
- Alexa, N.S.; James, E.P.; Terry, H.; Min, A. Identification and quantitation of compounds in a series of allelopathic and non-allelopathic rice root exudates. J. Chem. Ecol. 2004, 30, 1647–1662. [Google Scholar]
- Mattice, J.; Lavy, T.; Skulman, B.; Dilday, R.H. Searching for allelochemicals in rice that control ducksalad. In Allelopathy in Rice; Olofsdotter, M., Ed.; International Rice Research Institute: Manila, Philippines, 1998; pp. 154–196. [Google Scholar]
- Rice, E.L. Allelopathy—An update. Bot. Rev. 1979, 45, 15–109. [Google Scholar] [CrossRef]
- Inderjit. Soil Microorganisms: An important determinant of allelopathic activity. Plant Soil 2005, 274, 227–236. [Google Scholar] [CrossRef]
- Zhong, Y.G.; Chui, H.K.; Jing, G.W.; Yu, F. Rhizosphere isoflavones (daidzein and genistein) levels and their relation to the microbial community structure of mono-cropped soybean soil in field and controlled conditions. Soil Biol. Biochem. 2011, 43, 2257–2264. [Google Scholar]
- Dilday, R.H.; Nastasi, P.; Smith, R.J., Jr. Allelopathic observation in rice (Oryza sativa L.) to ducksalad (Heteranthera limosa). Proc. Ark. Acad. Sci. 1989, 43, 21–22. [Google Scholar]
- Chau, D.P.M.; Kieu, T.T.; Chin, D.V. Allelopathic effects of Vietnamese rice varieties. J. Allelopathy 2008, 22, 409–412. [Google Scholar]
- Khanh, T.D.; Xuan, T.D.; Chung, I.M. Rice allelopathy and the possibility for weed management. Ann. Appl. Biol. 2007, 151, 325–339. [Google Scholar] [CrossRef]
- Mew, T.W.; Alvarez, A.M.; Leach, J.E.; Swings, J. Focus on bacterial blight of rice. Plant Dis. 1993, 77, 5–12. [Google Scholar] [CrossRef]
- Chui, H.K.; Xiao, H.X.; Min, Z.; Song, Z.Z. Allelochemical tricin in rice hull and its aurone isomer against rice seedling rot disease. Pest Manag. Sci. 2010, 66, 1018–1024. [Google Scholar]
- Gun, W.L.; Moon, S.C.; Mihyung, K.; Byung, Y.C.; Sung, B.L. Direct suppression of a rice bacterial blight (Xanthomonas oryzae pv. oryzae) by monoterpene (S)-limonene. Protoplasma 2016, 253, 683–690. [Google Scholar]
- Morifumi, H.; Ichiro, M.; Shigemi, S.; Takuya, I.; Jinichiro, K.; Kazunori, O.; Hisakazu, Y.; Yuko, O. Phytoalexin accumulation in the interaction between rice and the blast fungus. Mol. Plant Microbe Interact. 2010, 23, 1000–1011. [Google Scholar]
- Yoshiki, K.; Jun, U.; Kimiko, K.; Yoshikatsu, S.; Masakazu, U.; Akira, S.; Minoru, W.; Tohru, T.; Daijiro, H.; Manabu, W.; et al. Structures of oryzalides A and B, and oryzalic acid A, a group of novel antimicrobial diterpenes, isolated from healthy leaves of a bacterial leaf blight-resistant cultivar of rice plant. Agric. Biol. Chem. 1991, 55, 803–811. [Google Scholar]
- Teresa, C.; Richard, M.T.; Victoria, A.N.; Julian, K.; Stefan, S.; Paul, C. Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci. 2020, 25, 635–643. [Google Scholar]
- Quan, N.V.; Dung, T.H.; Xuan, T.D.; Ateeque, A.; Dat, T.D.; Khanh, T.D.; Rolf, T. Momilactones A and B are α-amylase and α-glucosidase inhibitors. Molecules 2019, 24, 482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, N.V.; Dung, T.H.; Xuan, T.D.; Ateeque, A.; Dat, T.D.; Khanh, T.D. Contribution of momilactones A and B to diabetes inhibitory potential of rice bran: Evidence from in vitro assays. Saudi Pharm. J. 2019, 27, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Quan, N.V.; Thien, D.D.; Khanh, T.D.; Dung, T.H.; Xuan, T.D. Momilactones A, B, and tricin in rice grain and by-products are potential skin aging inhibitors. Foods 2019, 8, 602. [Google Scholar] [CrossRef] [PubMed]
- Pornngarm, L.; Supachai, Y.; Pornsiri, P.; Wanisa, P. Anti-inflammatory effects of proanthocyanidin-rich red rice extract via suppression of MAPK, AP-1 and NF-κB pathways in Raw 264.7 macrophages. Nutr. Res. Pract. 2016, 10, 251–258. [Google Scholar]
- Sun, J.K.; Hae, R.P.; Eunju, P.; Seung, C.L. Cytotoxic and antitumor activity of momilactone B from rice hulls. J. Agric. Food Chem. 2007, 55, 1702–1706. [Google Scholar]
- Chou, C.H.; Chang, F.J.; Oka, H.I. Allelopathic potentials of a wild rice, Oryza Perennis. Taiwania 1991, 36, 201–210. [Google Scholar]
- Salam, M.A.; Hisashi, K.N. Isolation and characterisation of two potent growth inhibitory substances from aqueous extract of Bangladeshi rice cultivar BR17. Allelopathy J. 2011, 27, 207–216. [Google Scholar]
- Hisashi, K.N.; Kazuya, T.; Hiroaki, S.; Kiyotake, S. Identification of two phytotoxins, blumenol A and grasshopper ketone, in the allelopathic Japanese rice variety Awaakamai. J. Plant Physiol. 2012, 169, 682–685. [Google Scholar]
- Xuan, T.D.; Minh, T.N.; Anh, L.H.; Khanh, T.D. Allelopathic momilactones A and B are implied in rice drought and salinity tolerance, not weed resistance. Agron. Sustain. Dev. 2016, 36, 52. [Google Scholar] [CrossRef]
- Chung, I.M.; Sung, K.P.; Mohd, A.; Mayakrishnan, P.; Young, T.O.; Seung, H.K.; Nasir, A.S.; Ateeque, A. Flavonoid glycosides from leaves and straw of Oryza sativa and their effects of cytotoxicity on a macrophage cell line and allelopathic on weed germination. Saudi Pharm. J. 2018, 26, 375–387. [Google Scholar] [CrossRef]
- Hisashi, K.N.; Morifumi, H.; Takeshi, I.; Katsumi, O.; Hiroya, K. Contribution of momilactone A and B to rice allelopathy. J. Plant Physiol. 2010, 167, 787–791. [Google Scholar]
- Ho, L.T.; Lin, C.H.; Smeda, R.J.; Fritschi, F.B. Isolation and purification of growth-inhibitors from Vietnamese rice cultivars. Weed Biol. Manag. 2014, 14, 221–231. [Google Scholar]
- Nguyen, C.; Vu, D.; Nguyen, N.Y.; Nguyen, T.T.T.; Phong, T.N.H.; Nguyen, C.T.; Lin, C.H.; Lei, Z.; Sumner, L.W. Allelopathic potential of rice and identification of published allelochemicals by cloud-based metabolomics platform. Metabolites 2020, 10, 244. [Google Scholar]
- Muller, C.H. The role of chemical inhibition (allelopathy) in vegetational composition. Bull. Torrey Bot. 1966, 99, 332–351. [Google Scholar] [CrossRef]
- Masakazu, F.; Xuan, T.D.; Farah, D.; Shinkichi, T.; Khanh, T.D.; Chung, I.M. Comparative efficacies in vitro of antibacterial, fungicidal, antioxidant, and herbicidal activities of momilatones A and B. J. Plant Interact. 2007, 2, 245–251. [Google Scholar]
- Bais, H.P.; Park, S.W.; Weir, T.L.; Callaway, R.M.; Vivanco, J.M. How plants communicate using the underground information superhighway. Trends Plant Sci. 2004, 9, 26–32. [Google Scholar] [CrossRef]
- Chung, I.M.; Ahn, J.K.; Yun, S.J. Identification of allelopathic compounds from rice (Oryza sativa L.) straw and their biological activity. Can. J. Plant Sci. 2001, 81, 815–819. [Google Scholar] [CrossRef]
- Shen, S.; Ma, G.; Xu, G.; Li, D.; Jin, G.; Yang, S.; Clements, D.R.; Chen, A.; Wen, L.; Zhang, F.; et al. Allelochemicals identified from sweet potato (Ipomoea batatas) and their allelopathic effects on invasive alien plants. Front. Plant Sci. 2022, 13, 823947. [Google Scholar] [CrossRef]
- Chandrasekara, A.; Shahidi, F. Bioactivities and antiradical properties of millet grains and hulls. J. Sci. Food Agric. 2011, 59, 9563–9571. [Google Scholar] [CrossRef]
- Tingting, Q.; Yong, S.; Xiaoya, W.; Liufeng, Z.; Hua, Z.; Li, J.; Xuemei, Z.; Hua, X. Drum drying-and extrusion-black rice anthocyanins exert anti-inflammatory effects via suppression of the NF-κB /MAPKs signaling pathways in LPS-induced RAW 264.7 cells. Food Biosci. 2020, 41, 100841. [Google Scholar]
- Ateeque, A.; Seung, H.K.; Mohd, A.; Inmyoung, P.; Jin, S.K.; Eun, H.K.; Ju, J.L.; Seul, K.K.; Chung, I.M. New chemical constituents from Oryza sativa straw and their algicidal activities against Blue-green algae. J. Agric. Food Chem. 2013, 61, 8039–8048. [Google Scholar]
- Osamu, K.; Junichi, M.; Tadami, A.; Shigehisa, K. Sakuranetin, a flavanone phytoalexin from ultraviolet-irradiated rice leaves. Phytochemistry 1992, 31, 3807–3809. [Google Scholar]
- Kong, C.H.; Zhao, H.; Xu, X.H.; Wang, P.; Gu, Y. Activity and allelopathy of soil of flavone O-Glycosides from rice. J. Agric. Food Chem. 2007, 55, 6007–6012. [Google Scholar] [CrossRef]
- Patterson, D.T. Effects of allelopathic chemicals on growth and physiological responses of soybean (Glycine max). Weed Sci. 1981, 29, 53–58. [Google Scholar] [CrossRef]
- Yoshida, S.; Asami, T.; Kawano, T.; Yoneyama, K.; Crow, W.D.; Paton, D.M.; Takahashi, N. Photosynthetic inhibitors in Eucalyptus grandis. Phytochemistry 1988, 27, 1943–1946. [Google Scholar] [CrossRef]
- Yu, J.Q.; Ye, S.F.; Zhang, M.F.; Hu, W.H. Effects of root exudates and aqueous root extracts of cucumber (Cucumis sativus) and allelochemicals, on photosynthesis and antioxidant enzymes in cucumber. Biochem. Syst. Ecol. 2003, 31, 129–139. [Google Scholar] [CrossRef]
- Zeng, R.S.; Luo, S.M.; Shi, Y.H. Physiological and biochemical mechanism of allelopathy of secalonic acid on higher plants. Agronomy 2001, 93, 72–79. [Google Scholar] [CrossRef]
- He, H.Q.; Lin, W.X. Studies on allelopathic physiobiochemical characteristics of rice. Chin. J. Eco-Agric. 2001, 9, 56–57. [Google Scholar]
- Chai, T.T.; Ooh, K.F.; Ooi, P.W.; Chue, P.S.; Wong, F.C. Leucaena leucocephala leachate compromised membrane integrity, respiration and antioxidative defence of water hyacinth leaf tissues. Bot. Stud. 2013, 54, 8. [Google Scholar] [CrossRef]
- Ye, S.F.; Zhou, Y.H.; Sun, Y.; Zou, L.Y.; Yu, J.Q. Cinnamic acid causes oxidative stress in cucumber roots, and promotes incidence of Fusarium wilt. Environ. Exp. Bot. 2006, 56, 255–262. [Google Scholar] [CrossRef]
- Shearer, T.L.; Rasher, D.B.; Snell, T.W.; Hay, M.E. Gene expression patterns of the coral Acropora millepora in response to contact with macroalgae. Coral Reefs 2012, 31, 1177–1192. [Google Scholar] [CrossRef] [PubMed]
- Li, H.H.; Nishimura, H.; Hasegawa, K.; Mizutani, J. Some physiological effects and the possible mechanism of action of juglone in plants. Weed Res. 1993, 38, 214–222. [Google Scholar] [CrossRef]
- Politycka, B. Free and glucosylated phenolics, phenol β-glucosyltransferase activity and membrane permeability in cucumber roots affected by derivatives of cinnamic and benzoic acids. Acta Physiol. Plant. 1997, 19, 311–317. [Google Scholar] [CrossRef]
- Kobza, J.; Einhellig, F.A. The effects of ferulic acid on the mineral nutrition of grain sorghum. Plant Soil. 1987, 98, 99–109. [Google Scholar] [CrossRef]
- Cheng, F.; Cheng, Z. Research progress on the use of plant allelopathy in agriculture and the physiological and ecological mechanisms of allelopathy. Front. Plant Sci. 2015, 6, 1020. [Google Scholar] [CrossRef]
- Wang, C.-M.; Chen, H.-T.; Li, T.-C.; Weng, J.-H.; Jhan, Y.-L.; Lin, S.-X.; Chou, C.-H. The role of pentacyclic triterpenoids in the allelopathic effects of Alstonia scholaris. J. Chem. Ecol. 2014, 40, 90–98. [Google Scholar] [CrossRef]
- Yoneyama, K.; Saruta, T.; Ogasawara, M.; Konnai, M.; Asami, T.; Abe, T.; Yoshida, S. Effects of grandinol and related phloroglucinol derivatives on transpiration and stomatal closure. Plant Growth Regul. 1996, 19, 7–11. [Google Scholar] [CrossRef]
- Anh, L.H.; Quan, N.V.; Nghia, L.T.; Xuan, T.D. Phenolic allelochemicals: Achievements, limitations, and prospective approaches in weed management. Weed Biol. Manag. 2021, 21, 37–67. [Google Scholar]
- Ni, H.W. Present status and prospect of crop allelopathy in China. In Rice Allelopathy; Kim, K.U., Shin, D.H., Eds.; Kyunpook National University: Kyungpook, Republic of Korea, 2000; pp. 41–48. [Google Scholar]
- Gomes, M.P.; Garcia, Q.S.; Barreto, L.C.; Pimenta, L.P.S.; Matheus, M.T.; Figueredo, C.C. Allelopathy: An overview from micro- to macroscopic organisms, from cells to environments, and the perspectives in a climate-changing world. Biologia 2017, 72, 113–129. [Google Scholar] [CrossRef]
- Mathesius, U.; Watt, M. Rhizosphere signals for plant—Microbe interactions: Implications for field—Grown plants. In Progress in Botany 72. Progress in Botany (Genetics—Physiology—Systematics—Ecology); Lüttge, U., Beyschlag, W., Büdel, B., Francis, D., Eds.; Springer: Berlin, Germany, 2010; pp. 125–161. [Google Scholar]
- Nardi, S.; Concheri, G.; Pizzeghello, D.; Sturaro, A.; Rella, R.; Parvoli, G. Soil organic matter mobilization by root exudates. Chemosphere 2000, 41, 653–658. [Google Scholar] [CrossRef]
- Macías, F.A.; Marín, D.; Oliveros-Bastidas, A.; Varela, R.M.; Simonet, A.M.; Carrera, C.; Molinillo, J.M. Allelopathy as a new strategy for sustainable ecosystems development. Biol. Sci. Space 2003, 17, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-P.; Feng, Y.-L.; Chen, Y.-J.; Tian, Y.-H. Soil microbes alleviate allelopathy of invasive plants. Sci. Bull. 2015, 60, 1083–1091. [Google Scholar] [CrossRef]
- Mishra, S.; Upadhyay, R.S.; Nautiyal, C.S. Unravelling the beneficial role of microbial contributors in reducing the allelopathic effects of weeds. Appl. Microbiol. Biotechnol. 2013, 97, 5659–5668. [Google Scholar] [CrossRef]
- Huang, S.H.; Ng, L.T. Quantification of polyphenolic content and bioactive constituents of some commercial rice varieties in Taiwan. J. Food Compos. Anal. 2012, 26, 122–127. [Google Scholar] [CrossRef]
- Er, S.G.; Shunjing, L.; Tong, L.; Chengmei, L.; Guowen, Z.; Jun, C.; Zicong, Z.; Rui, H. Phytochemical profiles and antioxidant activity of processed brown rice products. Food Chem. 2017, 232, 67–78. [Google Scholar]
- Chou, C.H. Autointoxication mechanism of Oryza sativa. I. Phytotoxic effects of decomposing rice residues in soils. J. Chem. Ecol. 1976, 2, 353–367. [Google Scholar] [CrossRef]
- Shicai, S.; Gaofeng, X.; David, R.C.; Guimei, J.; Fudou, Z.; Dayun, T.; Peng, X. Induced effects of exogenous phenolic acids on allelopathy of a wild rice accession (Oryza longistaminata, S37). Rice Sci. 2010, 17, 135–140. [Google Scholar]
- Quan, N.T.; Xuan, T.D. Foliar application of vanillic and p -hydroxybenzoic acids enhanced drought tolerance and formation of phytoalexin momilactones in rice. Arch. Agron. Soil Sci. 2018, 64, 1831–1846. [Google Scholar] [CrossRef]
- Xuan, T.D.; Bach, D.T.; Dat, T.D. Involvement of phenolics, flavonoids, and phenolic acids in high yield characteristics of rice (Oryza Sativa L.). Int. Lett. Nat. Sci. 2018, 68, 19–26. [Google Scholar] [CrossRef]
- Khanh, T.D.; Linh, L.H.; Linh, T.H.; Quan, N.T.; Cuong, D.M.; Hien, V.T.T.; Ham, L.H.; Xu, T.D. Integration of allelopathy to control weeds in rice. In Herbicides—Current Research and Case Studies in Use; Price, A.J., Kelton, J.A., Eds.; InTech: London, UK, 2013. [Google Scholar]
- Chen, J.; Yang, J.; Ma, L.; Li, J.; Shahzad, N.; Kim, C.K. Structure-antioxidant activity relationship of methoxyl, phenolic hydroxyl, and carboxylic acid groups of phenolic acids. Sci. Rep. 2020, 10, 2611. [Google Scholar] [CrossRef]
- Harukaze, A.; Murata, M.; Homma, S. Analyses of free and bound phenolics in rice. Food Sci. Technol. Res. 1999, 5, 74–79. [Google Scholar] [CrossRef]
- Hudson, E.A.; Dinh, P.A.; Kokubun, T.; Simmonds, M.S.; Gescher, A. Characterization of potentially chemopreventive phenols in extracts of brown rice that inhibit the growth of human breast and colon cancer cells. Cancer Epidemiol. Biomarkers Prev. 2000, 9, 1163–1170. [Google Scholar] [PubMed]
- Goufo, P.; Trindade, H. Rice antioxidants: Phenolic acids, flavonoids, anthocyanins, proanthocyanidins, tocopherols, tocotrienols, γ-oryzanol, and phytic acid. Food Sci. Nutr. 2014, 2, 75–104. [Google Scholar] [CrossRef] [PubMed]
- Seigler, D.S. Chemistry and mechanisms of allelopathic interactions. Agron. J. 1996, 88, 876–885. [Google Scholar] [CrossRef]
- Chung, I.M.; Ali, M.; Ateeque, A.; Jung, D.L.; Chang, Y.Y.; Jin, S.K. Chemical constituents of rice (Oryza sativa) hulls and their herbicidal activity against duckweed (Lemna paucicostata Hegelm 381). Phytochem. Anal. 2006, 17, 36–45. [Google Scholar] [CrossRef]
- Hooper, A.M.; Tsanuo, M.K.; Chamberlain, K.; Tittcomb, K.; Scholes, J.; Hassanali, A.; Khan, Z.R.; Pickett, J.A. Isoschaftoside, a C-glycosyl flavonoid from Desmodium uncinatum root exudate, is an allelochemical against the development of Striga. Phytochemistry 2010, 71, 904–908. [Google Scholar] [CrossRef]
- Mi, L.; Yunqiao, P.; Chang, G.Y.; Arthur, J.R. The occurrence of tricin and its derivatives in plants. Green Chem. 2016, 18, 1439–1454. [Google Scholar]
- Panyod, S.; Ho, C.-T.; Sheen, L.-Y. Dietary therapy and herbal medicine for COVID-19 prevention: A review and perspective. J. Tradit. Complement. Med. 2020, 10, 420–427. [Google Scholar] [CrossRef]
- Sen, S.; Chakraborty, R.; Kalita, P. Rice—Not just a staple food: A comprehensive review on its phytochemicals and therapeutic potential. Trends Food Sci. Technol. 2020, 97, 265–285. [Google Scholar] [CrossRef]
- Barbosa, J.R.; de Carvalho Junior, R.N. Polysaccharides obtained from natural edible sources and their role in modulating the immune system: Biologically active potential that can be exploited against COVID-19. Trends Food Sci. Technol. 2021, 108, 223–235. [Google Scholar] [CrossRef]
- Richard, D.V.; Peter, G.; Hong, C.; Arndt, B.; Massimo, B.; D’Incalci, M.; Riccio, E.; Rupa, D.; Izet, M.K.; William, P.S.; et al. Preliminary safety evaluation of the putative cancer chemopreventive agent tricin, a naturally occurring flavone. Cancer Chemother. Pharmacol. 2006, 57, 1–6. [Google Scholar]
- Smitha, M.; Rathnam, P.; Vasantha, S.; Antony, H.; Ananthasankaran, J. Isolation, characterization and quantification of tricin and flavonolignans in the medicinal rice Njavara (Oryza sativa L.), as compared to staple varieties. Plant Foods Hum. Nutr. 2011, 66, 91–96. [Google Scholar]
- Sompong, R.; Siebenhandl-Ehna, S.; Linsberger-Martina, G.; Berghofera, E. Physicochemical and antioxidative properties of red and black rice varieties from Thailand, China and Sri Lanka. Food Chem. 2011, 124, 132–140. [Google Scholar] [CrossRef]
- Sharifa, S.D.; Christine, B.; Siti, D.I.; Theja, H.; Robert, H.; Hueihong, L.; Fatemeh, H.; Priscila, Z.B.; Eduardo, G.; Julie, P.F.; et al. The potential of rice to offer solutions for malnutrition and chronic diseases. Rice 2012, 5, 16. [Google Scholar]
- Gema, P.C.; Shin, W.; Alan, C.; Tatsuhito, F.; Takao, Y.; Hiroshi, A. Phytochemical profile of a Japanese black–purple rice. Food Chem. 2013, 141, 2821–2827. [Google Scholar]
- Shao, Y.; Bao, J. Polyphenols in whole rice grain: Genetic diversity and health benefits. Food Chem. 2015, 180, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Yafang, S.; Feifei, X.; Xiao, S.; Jinsong, B.; Trust, B. Phenolic acids, anthocyanins, and antioxidant capacity in rice (Oryza sativa L.) grains at four stages of development after flowering. Food Chem. 2014, 143, 90–96. [Google Scholar]
- Pintha, K.; Yodkeeree, S.; Limtrakul, P. Proanthocyanidin in red rice inhibits MDA-MB-231 breast cancer cell invasion the expression control of invasive proteins. Biol. Pharm. Bull. 2015, 38, 571–581. [Google Scholar] [CrossRef]
- Mi, S.Y.; June, L.; Jin, M.L.; Younggyu, K.; Young, W.C.; Jun, G.J.; Young, S.K.; Yong, J.J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012, 22, 4049–4054. [Google Scholar]
- Hyman, B.T.; Hoesen, G.W.V.; Damasio, A.R.; Barnes, C.L. Alzheimer’s disease: Cell-specific pathology isolates the hippocampal formation. Science 1984, 225, 1168–1170. [Google Scholar] [CrossRef]
- Kato, H.; Kodama, O.; Akatsuka, T. Oryzalexin F, a diterpene phytoalexin from UV-irradiated rice leaves. Phytochemistry 1994, 36, 299–301. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Ino, T. Rice seedlings release momilactone B into the environment. Phytochemistry 2003, 63, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Quan, N.V.; Xuan, T.D.; Dung, T.H.; Thuy, N.T.D. Inhibitory activities of momilactones A, B, E, and 7-ketostigmasterol isolated from rice husk on paddy and invasive weeds. Plants 2019, 8, 159. [Google Scholar] [CrossRef]
- Akira, Y.; Naoto, S.; Osamu, K.; Tadami, A. Induction of phytoalexin formation in suspension-cultured rice cells by N-acetyl-chitooligosaccharides. Biosci. Biotechnol. Biochem. 1993, 57, 405–409. [Google Scholar]
- Minh, T.N.; Xuan, T.D.; Ateeque, A.; Abdelnaser, A.E.; Rolf, T.; Van, T.M. Momilactones A and B: Optimization of yields from isolation and purification. Separations 2018, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- Minh, T.N.; Xuan, T.D.; Ateeque, A.; Abdelnaser, A.E.; Rolf, T.; Van, T.M. Efficacy from different extractions for chemical profile and biological activities of rice husk. Sustainability 2018, 10, 1356. [Google Scholar] [CrossRef]
- Dong, Y.K.; Nipin, S.P.; Pramod, D.; Youn, H.Y.; Hyo, J.B.; Chang, H.D.; Kyung, D.P.; Mi, N.P.; Kwang, H.C.; Young, M.Y. Momilactone B inhibits ketosis in vitro by regulating the ANGPTL3-LPL pathway and inhibiting HMGCS2. Anim. Biotechnol. 2017, 28, 189–197. [Google Scholar]
- Keisuke, K.; Naoki, U.; Makoto, U.; Masayoshi, T.; Yutaka, O.; Naoki, M.; Kotomi, U.; Atsushi, I. Natural variation of diterpenoid phytoalexins in cultivated and wild rice species. Phytochemistry 2020, 180, 112518. [Google Scholar]
- Youn, H.J.; Eun, J.L.; Mi, S.K.; So, D.L.; So, Y.Y.; Young, C.L.; Young, B.Y.; Sang, K.Y.; Taekyu, P.; Chung, I.M.; et al. Enhancement of hypoxia-induced apoptosis of human breast cancer cells via STAT5b by momilactone B. Int. J. Oncol. 2008, 33, 477–484. [Google Scholar]
- Cheol, P.; Na, Y.J.; Gi, Y.K.; Min, H.H.; Chung, I.M.; Wun, J.K.; Young, H.Y.; Yung, H.C. Momilactone B induces apoptosis and G1 arrest of the cell cycle in human monocytic leukemia U937 cells through downregulation of pRB phosphorylation and induction of the cyclin-dependent kinase inhibitor p21Waf1/Cip1. Oncol. Rep. 2014, 31, 1653–1660. [Google Scholar]
- Chung, I.M.; Ali, M.; Hahn, S.J.; Siddiqui, N.A.; Lim, Y.H.; Ahmad, A. Chemical constituents from the hulls of Oryza sativa with cytotoxic activity. Chem. Nat. Compd. 2005, 41, 182–189. [Google Scholar] [CrossRef]
- Noriko, M.; Kotomi, U.; Masayoshi, T.; Yutaka, O.; Naoki, M.; Atsushi, I. Induced phenylamide accumulation in response to pathogen infection and hormone treatment in rice (Oryza sativa). Biosci. Biotechnol. Biochem. 2018, 82, 407–416. [Google Scholar]
- Ho, L.T.; Chung, H.L.; Reid, J.S.; Nathan, D.L.; Wei, G.W.; Felix, B.F. Isolation and identification of an allelopathic phenylethylamine in rice. Phytochemistry 2014, 108, 109–121. [Google Scholar]
- Tian, S.; Nakamura, K.; Kayahara, H. Analysis of phenolic compounds in white rice, brown rice, and germinated brown rice. J. Sci. Food Agric. 2004, 52, 4808–4813. [Google Scholar] [CrossRef] [PubMed]
- Olofsdotter, M. Allelopathy in Rice; International Rice Research Institute: Manila, Philippines, 1998; pp. 1–6. [Google Scholar]
- Agnes, M.R.; Maria, O.; Franck, E.D.; Stephen, O.D. Searching for rice allelochemicals. J. Agron. 2001, 93, 16–20. [Google Scholar]
- Francisco, A.M.; Nuria, C.; Rosa, M.V.; José, M.G.M. Bioactive steroids from Oryza sativa L. Steroids 2006, 71, 603–608. [Google Scholar]
- Yongxiang, L.; Hanyu, W.; Ruixue, Z.; Gang, S.; Songbo, W.; Ping, G.; Xiaotong, Z.; Qingyan, J.; Lina, W. Diet containing stearic acid increases food reward-related behaviors in mice compared with oleic acid. Brain Res. Bull. 2020, 164, 45–54. [Google Scholar]
- Takron, C.; Patinya, S.; Wichai, S.; Thawatchai, P. Vancomycin hydrochloride-loaded stearic acid/lauric acid in situ forming matrix for antimicrobial inhibition in patients with joint infection after total knee arthroplasty. Mater. Sci. Eng. C 2020, 115, 110761. [Google Scholar]
- Li, Y.; Kui, S.W.; Xiao, R.; Ying, X.Z.; Feng, W.; Qiang, W. Response of plant secondary metabolites to environmental factors. Molecules 2018, 23, 762. [Google Scholar]
- Butsat, S.; Weerapreeyakul, N.; Siriamornpun, S. Changes in phenolic acids and antioxidant activity in Thai rice husk at five growth stages during grain development. J. Agric. Food Chem. 2009, 57, 4566–4571. [Google Scholar] [CrossRef]
- Champagne, E.T.; Thompson, J.F.; Bett-Garber, K.L.; Mutters, R.; Miller, J.A.; Tan, E. Impact of storage of freshly harvested paddy rice on milled white rice flavor. Cereal Chem. 2004, 81, 444–449. [Google Scholar] [CrossRef]
- Bagchi, T.B.; Sharma, S.; Chattopadhyay, K. Development of NIRS models to predict protein and amylose content of brown rice and proximate compositions of rice bran. Food Chem. 2016, 191, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Zhongkai, Z.; Kevin, R.; Stuart, H.; Chris, B. The distribution of phenolic acids in rice. Food Chem. 2004, 87, 401–406. [Google Scholar]
- Fernanda, B.P.; Carolina, B.D.; Valéria, D.P.; Henrique, F.; Débora, L.M.; de Carvalho, L.M.; Carine, V.; Osmar, L.; Fernanda, O.L.; Lucas, B.; et al. Qualitative and quantitative analysis of the phenolic content of Connarus var. angustifolius, Cecropia obtusa, Cecropia palmata and Mansoa alliacea based on HPLC-DAD and UHPLC-ESI-MS/MS. Rev. Bras. Farmacogn. 2017, 27, 426–433. [Google Scholar]
- Steven, B.H.; Carol, B.G.; Kimberly, J.H.; David, J.M. Simple method for estimating polychlorinated biphenyl concentrations on soils and sediments using subcritical water extraction coupled with solid-phase microextraction. J. Chromatogr. A 1998, 814, 151–160. [Google Scholar]
- Hartonen, K.; Inkala, K.; Kangas, M.; Riekkola, M.L. Extraction of polychlorinated biphenyls with water under subcritical conditions. J. Chromatogr. A 1997, 785, 219–226. [Google Scholar] [CrossRef]
- Camel, V. Recent extraction techniques for solid matrices—Supercritical fluid extraction, pressurized fluid extraction and microwave-assisted extraction: Their potential and pitfalls. Analyst 2001, 126, 1182–1193. [Google Scholar] [CrossRef] [PubMed]
- Anne, B.; Serge, R.; Lidia, M.; Philippe, C.; Jean, L.V. Optimisation of accelerated solvent extraction of cocaine and benzoylecgonine from coca leaves. J. Sep. Sci. 2001, 24, 865–873. [Google Scholar]
- Eskilsson, C.S.; Sporring, S.; Björklund, E. Fast and selective analytical procedures for determination of persistent organic pollutants in food and feed using recent extraction techniques in modern extraction techniques. J. Am. Chem. Soc. 2006, 26, 145. [Google Scholar]
- Camel, V. Microwave-assisted solvent extraction of environmental samples. TrAC Trend. Anal. Chem. 2000, 19, 229–248. [Google Scholar] [CrossRef]
- Letellier, M.; Budzinski, H. Microwave assisted extraction of organic compounds. Analysis 1999, 27, 259–270. [Google Scholar] [CrossRef]
- Hostettmann, K.; Marston, A.; Hostettmann, M. Preparative Chromatography Techniques: Applications in Natural Product Isolation, 2nd ed.; Springer: Berlin, Germany, 2010. [Google Scholar]
- Sturm, S.; Seger, C. Liquid chromatography–nuclear magnetic resonance coupling as alternative to liquid chromatography–mass spectrometry hyphenations: Curious option or powerful and complementary routine tool. J. Chromatogr. A 2012, 1259, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Prosen, H.; Zupančič-Kralj, L. Solid-phase microextraction. Trends Analyt. Chem. 1999, 18, 272–282. [Google Scholar] [CrossRef]
- Widiastuti, S.; Irfan, E.S.; Ceferino, A.C.; Miguel, P. Optimisation of an ultrasound-assisted extraction method for the simultaneous determination of phenolics in rice grains. Food Chem. 2019, 288, 221–227. [Google Scholar]
- Setyaningsih, W.; Saputro, I.E.; Palma, M.; Barroso, C.G. Pressurized liquid extraction of phenolic compounds from rice (Oryza sativa) grains. Food Chem. 2016, 192, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Setyaningsiha, W.; Saputro, I.E.; Palma, M.; Barroso, C.G. Optimisation and validation of the microwave-assisted extraction of phenolic compounds from rice grains. Food Chem. 2015, 169, 141–149. [Google Scholar] [CrossRef]
- Harish, C.A.; Vijay, K.P. High performance thin layer chromatography (HPTLC): A modern analytical tool for biological analysis. Nat. Sci. 2010, 8, 58–61. [Google Scholar]
- Monika, S.; Sukanya, P.; Rajesh, K.S. Common techniques and methods for screening of natural products for developing of anticancer drugs. In Evolutionary Diversity as a Source for Anticancer Molecules; Srivastava, A.K., Kannaujiya, V.K., Singh, R.K., Singh, D., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 323–353. [Google Scholar]
- Reversed Phase Chromatography: Principles and Methods. Available online: http://wolfson.huji.ac.il/purification/PDF/ReversePhase/AmershamRPCManual.pdf (accessed on 12 October 2022).
- Kemp, W. Energy and electromagnetic spectrum. In Organic Spectroscopy; Kemp, W., Ed.; Macmillan Press: London, UK, 1991; pp. 1–7. [Google Scholar]
- Urbano, M.; Luque de Castro, M.D.; Pérez, P.M.; García-Olmo, J.; Gómez-Nieto, M.A. Ultraviolet–visible spectroscopy and pattern recognition methods for differentiation and classification of wines. Food Chem. 2006, 97, 166–175. [Google Scholar] [CrossRef]
- Larissa, N.R.; Aline, C.; Paulo, H.M.; Patrícia, V. Thermal rice oil degradation evaluated by UV–Vis-NIR and PARAFAC. Food Chem. 2019, 273, 52–56. [Google Scholar]
- Fang, X.; Yu, X.; Rui, L.; Guo, P.S.; Han, Q.Y. Elucidation of the thermal deterioration mechanism of bio-oil pyrolyzed from rice husk using fourier transform infrared spectroscopy. J. Agric. Food Chem. 2011, 59, 9243–9249. [Google Scholar]
- Beckey, H.D. 1–Theory of Field Ionization (FI) and Field Emission (FE). In Field Ionization Mass Spectrometry; Beckey, H.D., Ed.; Pergamon: Bergama, Turkey, 1971. [Google Scholar]
- Beckey, H.D. 2–Field Ionization Sources. In Field Ionization Mass Spectrometry; Beckey, H.D., Ed.; Pergamon: Bergama, Turkey, 1971. [Google Scholar]
- Beckey, H.D. 3–Application of the FI Mass Spectrometer to Physico-Chemical Problems. In Field Ionization Mass Spectrometry; Beckey, H.D., Ed.; Pergamon: Bergama, Turkey, 1971. [Google Scholar]
- Beckey, H.D. 4–Qualitative Analyses with the Fi Mass Spectrometer. In Field Ionization Mass Spectrometry; Beckey, H.D., Ed.; Pergamon: Bergama, Turkey, 1971. [Google Scholar]
- Mukherjee, P.K. Bioassay-guided isolation and evaluation of herbal drugs. In Evaluating Natural Products and Traditional Medicine; Mukherjee, P.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 515–537. [Google Scholar]
- Susana, P.G.; Florbela, P. Dereplication: Racing to speed up the natural products discovery process. Nat. Prod. Rep. 2015, 32, 779–810. [Google Scholar]
- Sherma, J.; Fried, B. Handbook of Thin-Layer Chromatography; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Waksmundzka-Hajnos, M.; Sherma, J. High Performance Liquid Chromatography in Phytochemical Analysis; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Wolfender, J.-L. HPLC in natural product analysis: The detection Issue. Planta. Med. 2009, 75, 719–734. [Google Scholar] [CrossRef]
- Herrero, M.; Castro-Puyana, M.; Ibáñez, E.; Cifuentes, A. Compositional analysis of foods. In Liquid Chromatography; Fanali, S., Haddad, P., Poole, C., Schoenmakers, P., Lloyd, D., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 295–317. [Google Scholar]
- Chatthongpisut, R.; Schwartz, S.J.; Yongsawatdigul, J. Antioxidant activities and antiproliferative activity of Thai purple rice cooked by various methods on human colon cancer cells. Food Chem. 2015, 188, 99–105. [Google Scholar] [CrossRef]
- Jandera, P. Stationary and mobile phases in hydrophilic interaction chromatography: A review. Anal. Chim. Acta 2011, 692, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.C.; Xin, M.L.; Ning, Y.; Feng, Z.W.; Yong, M.D.; Zhong, F.Z. Partial purification, identification, and quantitation of antioxidants from wild rice (Zizania latifolia). Molecules 2018, 23, 2782. [Google Scholar]
- Meixue, Z.; Kevin, R.; Malcolm, G.H.; Stuart, H. Analysis of volatile compounds and their contribution to flavor in Cereals. J. Agric. Food Chem. 1999, 47, 3941–3953. [Google Scholar]
- Delahunty, C.M.; Eyres, G.; Dufour, J.-P. Gas chromatography-olfactometry. J. Sep. Sci. 2006, 29, 2107–2125. [Google Scholar] [CrossRef] [PubMed]
- Manabu, W.; Yoshiki, K.; Yasuaki, E.; Tohru, T.; Daijiro, H.; Yoshikatsu, S.; Akira, S.; Minoru, W. Studies on a quantitative analysis of oryzalides and oryzalic acids in rice plants by GC-SIM. Biosci. Biotechnol. Biochem. 1996, 60, 1460–1463. [Google Scholar]
- Kong, C.H.; Wang, P.; Zhao, H.; Xu, X.H.; Zhu, Y.D. Impact of allelochemical exuded from allelopathic rice on soil microbial community. Soil Biol. Biochem. 2008, 40, 1862–1869. [Google Scholar] [CrossRef]
- Gross, J.H. Mass spectrometry: A Textbook; Springer: Berlin, Germany, 2004. [Google Scholar]
- Das, A.B.; Goud, V.V.; Das, C. Extraction of phenolic compounds and anthocyanin from black and purple rice bran (Oryza sativa L.) using ultrasound: A comparative analysis and phytochemical profiling. Ind. Crops Prod. 2017, 95, 332–341. [Google Scholar] [CrossRef]
- You, L.X.; Wang, P.; Kong, C.H. The levels of jasmonic acid and salicylic acid in a rice-barnyardgrass coexistence system and their relation to rice allelochemicals. Biochem. Syst. Ecol. 2011, 39, 491–497. [Google Scholar] [CrossRef]
- Jung, Y.H.; Lee, J.H.; Agrawal, G.K.; Rakwal, R.; Kim, J.A.; Shim, J.K.; Lee, S.K.; Jeon, J.S.; Koh, H.J.; Lee, Y.H.; et al. The rice (Oryza sativa) blast lesion mimic mutant, blm, may confer resistance to blast pathogens by triggering multiple defense-associated signaling pathways. Plant Physiol. Biochem. 2005, 43, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Flávia, S.S.; Clarissa, O.S.; Vivian, C.B.; Ana, P.B.F.; Marcela, B.S.; Andréa, C.D.; José, L.N. Analysis of polyphenols in brewer’s spent grain and its comparison with corn silage and cereal brans commonly used for animal nutrition. Food Chem. 2018, 239, 385–401. [Google Scholar]
- DeVierno, K.A.; House, K.T.; Whitford, J.; Ponnusamy, E.; Miller, P.; Jesse, N.; Rodenborn, R.; Sayag, S.; Gebel, M.; Aped, I.; et al. A method for assessing greener alternatives between chemical Products following the 12 principles of green chemistry. ACS Sustain. Chem. Eng. 2017, 5, 2927–2935. [Google Scholar] [CrossRef]
- Singh, A.; Rajeswari, G.; Nirmal, L.; Jacob, S. Synthesis and extraction routes of allelochemicals from plants and microbes: A review. Rev. Anal. Chem. 2021, 40, 293–311. [Google Scholar] [CrossRef]
- Khoddami, A.; Wilkes, M.A.; Roberts, T.H. Techniques for analysis of plant phenolic compounds. Molecules 2013, 18, 2328–2375. [Google Scholar] [CrossRef]
- Lindquist, E.; Yang, Y. Degradation of benzoic acid and its derivatives in subcritical water. J. Chromatogr. A 2011, 1218, 2146–2152. [Google Scholar] [CrossRef]
- Cravottoa, G.; Boffaa, L.; Mantegnaa, S.; Peregob, P.; Avogadrob, M.; Cintasc, P. Improved extraction of vegetable oils under high-intensity ultrasound and/or microwaves. Ultrason. Sonochem. 2008, 15, 898–902. [Google Scholar] [CrossRef]









| Modes of Action | Allelochemicals | Recipient Plant | Rice Organs | References |
|---|---|---|---|---|
| Growth inhibition | ||||
| Inhibition of root and hypocotyl growth | Momilactone A (MA) and B (MB) | Lepidium sativum seedlings, Lactuca sativa seedlings, Echinochloa crus-galli, and Monochoria vaginalis | Husks, leaves, brans, roots, and root exudates | [5,38] |
| Root growth suppression | Gallic acid, protocatechuic acid, p-hydroxybenzoic, vanillic acid, syringic acid, p-coumaric acid, m-coumaric acid, ferulic acid, o-coumaric acid | Brassica rapa and O. sativa seeds. | Leaves and stems | [29] |
| Inhibition of root and shoot growth | Blumenol A and grasshopper ketone | L. sativum, L. sativa, Phleum pratense, Digitaria sanguinalis, Lolium multiflorum, and E. crus-galli. | Whole plant | [31] |
| Inhibition of the radicle growth | p-Hydroxybenzoic, p-coumaric, vanillic, ferulic, and o-hydroxyphenylacetic acids, and several unknowns | L. sativa seeds, O. sativa seeds and seedlings. | Residues in soil | [39] |
| Inhibition of seed germination, seedling length, and dry weight | p-Hydroxybenzoic acid, p-coumaric acid, ferulic acid, and p-hydroxybenzoic acid | E. crus-galli | Straws | [40] |
| Inhibition of plant height, root length, monocotyledon and fresh weight | Salicylic acid, fumaric acid, p-coumaric acid and p-hydroxybenzonic acid | E. crus-galli | Whole plant | [41] |
| - | Lanast-7,9(11)-dien-3α,15α-diol-3α-d-glucofuranoside | Lemna paucicostata | Hulls | [42] |
| Inhibition of germination rate, seedling growth, shoots, and roots | Momilactone E (ME), 7-ketostigmasterol (7KS), MA, and MB | L. sativa, E. crus-galli, and Solidago altissima | Husks | [43] |
| Algicidal activities | Oleioyl-β-d-arabinoside | Cyanobacteria | Straws | [44] |
| Inhibition of spore germination | Sakuranetin | Pyricularia oryzae | Leaves | [45] |
| - | Two flavone O-glycosides | Interferes with weeds or microbes in paddy soil. | Seedlings | [46] |
| Physiological pathways | ||||
| Inhibition of photosynthesis | Caffeic, coumaric, ferulic, cinnamic, and vanillic acids | - | - | [47,48] |
| Suppress respiration | Benzoic and cinnamic acids | - | - | [49] |
| Enzyme functions | ||||
| Phosphorylase suppressors | Chlorogenic, caffeic acids, and catechol | - | - | [50,51] |
| ATPase inhibitors | Cinnamic acid and its derivatives | E. crus-galli | Whole plant | [50,51] |
| Rhizosphere microorganisms | ||||
| Suppression of nitrification process, by inhibiting the activities of vital enzymes such as ammonium mono-oxygenase and hydroxylamine oxidoreductase | Methyl 3-(4-hydroxyphenyl) propionate, linoleic acid, methyl-p-coumarate and methyl ferulate | - | Whole plant, root exudates | [6] |
| Inhibition of the oxidation of NH 4+ to NO2- | Caffeic and ferulic acids, myricetin, tannins and tannin derivatives | - | - | [12] |
| Synergistic suppressive effect by reducing nitrogen-fixing ability | Different phenolic compounds | Rhizobium strain | Decomposition of rice residue and straw in soil | [6] |
| Affecting soil community structure and reducing fungi present in paddy soil | Tricin (5,7,4′-trihydroxy-3′,5′-dimethoxyflavone) and aurone isomer (5,7,4′-trihydroxy-3′,5′-dimethoxyaurone) of tricin | Fusarium oxysporum and Rhizoctonia solani | Hulls, leaves, roots, and root exudates | [19,46] |
| Pathogen infection | ||||
| Suppressing Xoo growth in rice seedlings | (S)-Limonene, oryzalides A and B, and oryzalic acid A | Xanthomonas oryzae | Seedlings and leaves | [20,22] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anh, N.T.H.; Anh, L.H.; Mai, N.P.; Quan, N.V.; Xuan, T.D. Identification and Isolation Techniques for Plant Growth Inhibitors in Rice. Separations 2023, 10, 105. https://doi.org/10.3390/separations10020105
Anh NTH, Anh LH, Mai NP, Quan NV, Xuan TD. Identification and Isolation Techniques for Plant Growth Inhibitors in Rice. Separations. 2023; 10(2):105. https://doi.org/10.3390/separations10020105
Chicago/Turabian StyleAnh, Nguyen Thi Hai, La Hoang Anh, Nguyen Phuong Mai, Nguyen Van Quan, and Tran Dang Xuan. 2023. "Identification and Isolation Techniques for Plant Growth Inhibitors in Rice" Separations 10, no. 2: 105. https://doi.org/10.3390/separations10020105
APA StyleAnh, N. T. H., Anh, L. H., Mai, N. P., Quan, N. V., & Xuan, T. D. (2023). Identification and Isolation Techniques for Plant Growth Inhibitors in Rice. Separations, 10(2), 105. https://doi.org/10.3390/separations10020105