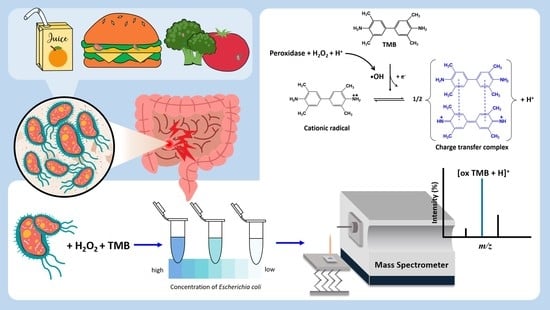
Detection of Escherichia coli by Combining an Affinity-Based Method with Contactless Atmospheric Pressure Ionization Mass Spectrometry
Abstract
:1. Introduction
2. Experimental Section
2.1. Bacterial Endogenous Peroxidase Reactions
2.2. Preparation of Affinity Probes
2.3. Bacterial Enrichment and Endogenous Peroxidase Reactions
2.4. C-API MS Analysis
2.5. Analysis of Simulated Real Samples
3. Results and Discussion
3.1. Characterization of Affinity Probes
3.2. Direct Analysis of the Products Derived from Horseradish Peroxidase Enzymatic Reaction by C-API-MS
3.3. Direct Analysis of Bacterial Endogenous Peroxidase-Based Reaction Products by C-API-MS
3.4. Enrichment of Target Bacteria by the MALCY-Probe
3.5. Examination of LOD
3.6. Analysis of Different E. coli Strains
3.7. Examination of Selectivity
3.8. Analysis of Bacteria in Simulated Real Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fung, F.; Wang, H.-S.; Menon, S. Food safety in the 21st century. Biomed. J. 2018, 41, 88–95. [Google Scholar]
- Motarjemi, Y.; Käferstein, F.; Moy, G.; Quevedo, F. Contaminated weaning food: A major risk factor for diarrhoea and associated malnutrition. Bull. WHO 1993, 71, 79–92. [Google Scholar]
- Tarr, P.I.; Neill, M.A. Escherichia coli O157:H7. Gastroenterol. Clin. North Am. 2001, 30, 735–751. [Google Scholar] [CrossRef]
- Butler, D. Novel pathogens beat food safety checks. Nature 1996, 384, 397. [Google Scholar] [CrossRef]
- Nguyen, Y.; Sperandio, V. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front. Cell. Infect. Microbiol. 2012, 2, 90. [Google Scholar] [CrossRef] [Green Version]
- Eum, N.-S.; Yeom, S.-H.; Kwon, D.-H.; Kim, H.-R.; Kang, S.-W. Enhancement of sensitivity using gold nanorods—Antibody conjugator for detection of E. coli O157:H7. Sens. Actuators B 2010, 143, 784–788. [Google Scholar] [CrossRef]
- Fu, Z.; Rogelj, S.; Kieft, T.L. Rapid detection of Escherichia coli O157:H7 by immunomagnetic separation and real-time PCR. Int. J. Food Microbiol. 2005, 99, 47–57. [Google Scholar] [CrossRef]
- Daly, P.; Collier, T.; Doyle, S. PCR-ELISA detection of Escherichia coli in milk. Lett. Appl. Microbiol. 2002, 34, 222–226. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zeng, D.; Yan, C.; Chen, W.; Ren, J.; Jiang, Y.; Jiang, L.; Xue, F.; Ji, D.; Tang, F.; et al. Rapid and accurate detection of Escherichia coli O157:H7 in beef using microfluidic wax-printed paper-based ELISA. Analyst 2020, 145, 3106–3115. [Google Scholar] [CrossRef]
- Jin, D.; Qi, H.; Chen, S.; Zeng, T.; Liu, Q.; Wang, S. Simultaneous detection of six human diarrheal pathogens by using DNA microarray combined with tyramide signal amplification. J. Microbiol. Methods 2008, 75, 365–368. [Google Scholar] [CrossRef]
- Borst, A.; Box, A.T.A.; Fluit, A.C. False-Positive Results and Contamination in Nucleic Acid Amplification Assays: Suggestions for a Prevent and Destroy Strategy. Eur. J. Clin. Microbiol. Infect. Dis. 2004, 23, 289–299. [Google Scholar] [CrossRef]
- Wolter, A.; Niessner, R.; Seidel, M. Detection of Escherichia coli O157:H7, Salmonella typhimurium, and Legionella pneumophila in Water Using a Flow-Through Chemiluminescence Microarray Readout System. Anal. Chem. 2008, 80, 5854–5863. [Google Scholar] [CrossRef]
- Gupta, R.; Kumar, A.; Kumar, S.; Pinnaka, A.K.; Singhal, N.K. Naked eye colorimetric detection of Escherichia coli using aptamer conjugated graphene oxide enclosed Gold nanoparticles. Sens. Actuators B 2021, 329, 129100. [Google Scholar] [CrossRef]
- Ahmad, M.; Roberts, J.N.; Hardiman, E.M.; Singh, R.; Eltis, L.D.; Bugg, T.D.H. Identification of DypB from Rhodococcus jostii RHA1 as a Lignin Peroxidase. Biochemistry 2011, 50, 5096–5107. [Google Scholar] [CrossRef]
- Liu, L.; Hao, Y.; Deng, D.; Xia, N. Nanomaterials-Based Colorimetric Immunoassays. Nanomaterials 2019, 9, 316. [Google Scholar] [CrossRef] [Green Version]
- Josephy, P.D.; Eling, T.; Mason, R.P. The horseradish peroxidase-catalyzed oxidation of 3, 5, 3′, 5′-tetramethylbenzidine. Free radical and charge-transfer complex intermediates. J. Biol. Chem. 1982, 257, 3669–3675. [Google Scholar] [CrossRef]
- Huang, H.; Wu, Z.; Huang, J.; Zhao, G.; Dou, W. Highly sensitive colorimetric immunoassay for Escherichia coli O157:H7 based on probe of pseudo enzyme and dual signal amplification. Anal. Methods. 2018, 10, 4301–4309. [Google Scholar] [CrossRef]
- Shim, K.H.; Kang, M.; Kim, M.G.; Chung, B.H.; An, S.S.A. Detection of E. coli O157:H7 using its endogenous active membrane peroxidase. J. Toxicol. Environ. Health Sci. 2011, 3, 80–85. [Google Scholar] [CrossRef]
- Su, H.; Zhao, H.; Qiao, F.; Chen, L.; Duan, R.; Ai, S. Colorimetric detection of Escherichia coli O157:H7 using functionalized Au@Pt nanoparticles as peroxidase mimetics. Analyst 2013, 138, 3026–3031. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, K.; Li, N.; He, J.; Qiao, L.; Li, M.; Wang, Y.; Zhang, D.; Jiang, J.; Zare, R.N. Real-time mass-spectrometric screening of droplet-scale electrochemical reactions. Analyst 2018, 143, 4247–4250. [Google Scholar] [CrossRef]
- Mai-Prochnow, A.; Lucas-Elio, P.; Egan, S.; Thomas, T.; Webb, J.S.; Sanchez-Amat, A.; Kjelleberg, S. Hydrogen Peroxide Linked to Lysine Oxidase Activity Facilitates Biofilm Differentiation and Dispersal in Several Gram-Negative Bacteria. J. Bacteriol. 2008, 190, 5493–5501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nóbrega, C.S.; Pauleta, S.R. Reduction of hydrogen peroxide in gram-negative bacteria—Bacterial peroxidases. Adv. Microb. Physiol. 2019, 74, 415–464. [Google Scholar] [CrossRef] [PubMed]
- Pieters, R.J. Maximising multivalency effects in protein–carbohydrate interactions. Org. Biomol. Chem. 2009, 7, 2013–2025. [Google Scholar] [CrossRef]
- Qiao, Z.; Lei, C.; Fu, Y.; Li, Y. An antimicrobial peptide-based colorimetric bioassay for rapid and sensitive detection of E. coli O157:H7. RSC Adv. 2017, 7, 15769–15775. [Google Scholar] [CrossRef]
- Asadi, A.; Razavi, S.; Talebi, M.; Gholami, M. A review on anti-adhesion therapies of bacterial diseases. Infection 2019, 47, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Biscola, F.T.; Abe, C.M.; Guth, B.E.C. Determination of adhesin gene sequences in, and biofilm formation by, O157 and non-O157 Shiga toxin-producing Escherichia coli strains isolated from different sources. Appl. Environ. Microbiol. 2011, 77, 2201–2208. [Google Scholar] [CrossRef] [Green Version]
- Miller, E.; Garcia, T.; Hultgren, S.; Oberhauser, A.F. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys. J. 2006, 91, 3848–3856. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, J.; Berglund, J.; Schembri, M.; De Genst, E.; Cools, L.; Wuhrer, M.; Hung, C.S.; Pinkner, J.; Slattegard, R.; Zavialov, A.; et al. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol. Microbiol. 2005, 55, 441–455. [Google Scholar] [CrossRef]
- Lundquist, J.J.; Toone, E.J. The Cluster Glycoside Effect. Chem. Rev. 2002, 102, 555–578. [Google Scholar] [CrossRef]
- Yan, X.; Sivignon, A.; Yamakawa, N.; Crepet, A.; Travelet, C.; Borsali, R.; Dumych, T.; Li, Z.; Bilyy, R.; Deniaud, D.; et al. Glycopolymers as Antiadhesives of E. coli Strains Inducing Inflammatory Bowel Diseases. Biomacromolecules 2015, 16, 1827–1836. [Google Scholar] [CrossRef]
- Kang, T.W.; Han, J.; Lee, S.; Hwang, I.-J.; Jeon, S.-J.; Ju, J.-M.; Kim, M.-J.; Yang, J.-K.; Jun, B.; Lee, C.H.; et al. 2D transition metal dichalcogenides with glucan multivalency for antibody-free pathogen recognition. Nat. Commun. 2018, 9, 2549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, C.H.; Chang, C.H.; Urban, P.L.; Chen, Y.C. Capillary action-supported contactless atmospheric pressure ionization for the combined sampling and mass spectrometric analysis of biomolecules. Anal. Chem. 2011, 83, 2866–2869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, C.-H.; Meher, A.K.; Chen, Y.-C. Automatic sampling and analysis of organics and biomolecules by capillary action-supported contactless atmospheric pressure ionization mass spectrometry. PLoS ONE 2013, 8, e66292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-C.; Krishnamurthy, A.; Chen, S.-H.; Chen, Y.-C. A tapered capillary-based contactless atmospheric pressure ionization mass spectrometry for on-line preconcentration and separation of small organics. Separations 2021, 8, 111. [Google Scholar] [CrossRef]
- Li, L.; Liao, L.; Ding, Y.; Zeng, H. Dithizone-etched CdTe nanoparticles-based fluorescence sensor for the off–on detection of cadmium ion in aqueous media. RSC Adv. 2017, 7, 10361–10368. [Google Scholar] [CrossRef] [Green Version]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.-I.; Lee, Y.C. Carbohydrate analysis by a phenol–sulfuric acid method in microplate format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef]
- Goodridge, L.; Chen, J.; Griffiths, M. Development and Characterization of a Fluorescent-Bacteriophage Assay for Detection of Escherichia coli O157:H7. Appl. Environ. Microbiol. 1999, 65, 1397–1404. [Google Scholar] [CrossRef] [Green Version]
- Ye, L.; Zhao, G.; Dou, W. An electrochemical immunoassay for Escherichia coli O157:H7 using double functionalized Au@Pt/SiO2 nanocomposites and immune magnetic nanoparticles. Talanta 2018, 182, 354–362. [Google Scholar] [CrossRef]
- Ramasamy, M.; Yi, D.K.; An, S.S.A. Enhanced detection sensitivity of Escherichia coli O157:H7 using surface-modified gold nanorods. Int. J. Nanomed. 2015, 10, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Qiao, L.; Prudent, M.; Bondarenko, A.; Gasilova, N.; Möller, S.B.; Lion, N.; Pick, H.; Gong, T.; Chen, Z.; et al. Sensitive and fast identification of bacteria in blood samples by immunoaffinity mass spectrometry for quick BSI diagnosis. Chem. Sci. 2016, 7, 2987–2995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, R.; Cao, Z.; Li, H.; Graw, J.; Zhang, G.; Thannickal, V.J.; Cheng, G. Peroxidasin contributes to lung host defense by direct binding and killing of gram-negative bacteria. PLoS Pathog. 2018, 14, e1007026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustafa, H.S.I. Staphylococcus aureus can produce catalase enzyme when adding to human wbcs as a source of H2O2 productions in human plasma or serum in the laboratory. Open J. Med. Microbiol. 2014, 4, 249. [Google Scholar] [CrossRef] [Green Version]
- Shmaefsky, B.R. Artificial urine for laboratory testing. Am. Biol. Teach. 1990, 52, 170–172. [Google Scholar] [CrossRef]
- Johnson, J.R.; Russo, T.A.; Scheutz, F.; Brown, J.J.; Zhang, L.; Palin, K.; Rode, C.; Bloch, C.; Marrs, C.F.; Foxman, B. Discovery of disseminated J96-like strains of uropathogenic Escherichia coli O4:H5 containing genes for both PapG(J96) (class I) and PrsG(J96) (class III) Gal(alpha1-4)Gal-binding adhesins. J. Infect. Dis. 1997, 175, 983–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sari, J.N.; Kandasamy, K.; Chen, Y.-C. Detection of Escherichia coli by Combining an Affinity-Based Method with Contactless Atmospheric Pressure Ionization Mass Spectrometry. Separations 2022, 9, 13. https://doi.org/10.3390/separations9010013
Sari JN, Kandasamy K, Chen Y-C. Detection of Escherichia coli by Combining an Affinity-Based Method with Contactless Atmospheric Pressure Ionization Mass Spectrometry. Separations. 2022; 9(1):13. https://doi.org/10.3390/separations9010013
Chicago/Turabian StyleSari, Juli Novita, Karthikeyan Kandasamy, and Yu-Chie Chen. 2022. "Detection of Escherichia coli by Combining an Affinity-Based Method with Contactless Atmospheric Pressure Ionization Mass Spectrometry" Separations 9, no. 1: 13. https://doi.org/10.3390/separations9010013
APA StyleSari, J. N., Kandasamy, K., & Chen, Y. -C. (2022). Detection of Escherichia coli by Combining an Affinity-Based Method with Contactless Atmospheric Pressure Ionization Mass Spectrometry. Separations, 9(1), 13. https://doi.org/10.3390/separations9010013