Circulating Tumor Cell Models Mimicking Metastasizing Cells In Vitro: Discrimination of Colorectal Cancer Cells and White Blood Cells Using Digital Holographic Cytometry
Abstract
:1. Introduction
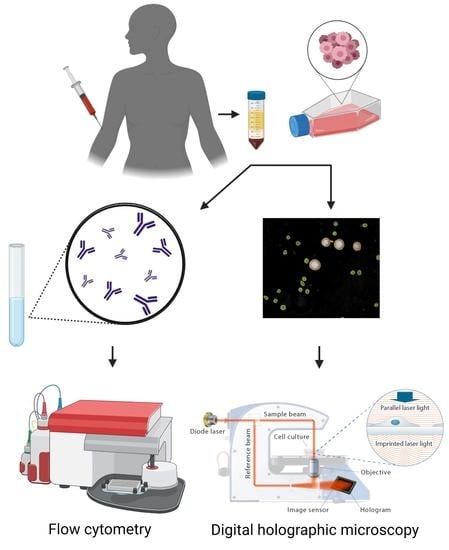
2. Materials and Methods
2.1. Cell Culture
2.2. Isolation of Peripheral Blood Mononuclear Cells (PBMCs)
2.3. Fluorescence-Activated Cell Sorting
2.4. DHC and Image Analysis
2.5. Statistics
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.H.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis. Primers 2015, 1, 15065. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA A Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, R.A.; Asmis, T.R. Overview of systemic therapy for colorectal cancer. Clin. Colon Rectal Surg. 2009, 22, 251–256. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Kuipers, E.J.; Rösch, T.; Bretthauer, M. Colorectal cancer screening—Optimizing current strategies and new directions. Nat. Rev. Clin. Oncol. 2013, 10, 130–142. [Google Scholar] [CrossRef]
- Geiger, T.M.; Ricciardi, R. Screening options and recommendations for colorectal cancer. Clin. Colon Rectal Surg. 2009, 22, 209–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donato, C.; Kunz, L.; Castro-Giner, F.; Paasinen-Sohns, A.; Strittmatter, K.; Szczerba, B.M.; Scherrer, R.; Di Maggio, N.; Heusermann, W.; Biehlmaier, O.; et al. Hypoxia Triggers the Intravasation of Clustered Circulating Tumor Cells. Cell Rep. 2020, 32, 108105. [Google Scholar] [CrossRef]
- Massagué, J.; Obenauf, A. Metastatic colonization by circulating tumor cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Weinberg, R.A. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015, 25, 675–686. [Google Scholar] [CrossRef] [Green Version]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Clark, A.G.; Vignjevic, D.M. Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 2015, 36, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Gjörloff-Wingren, A. Quantitative phase-contrast imaging—A potential tool for future cancer diagnostics. Cytom. Part A 2017, 91, 752–753. [Google Scholar] [CrossRef] [PubMed]
- Janicke, B.; Kårsnäs, A.; Egelberg, P.; Alm, K. Label-free high temporal resolution assessment of cell proliferation using digital holographic microscopy. Cytom. Part A 2017, 91, 460–469. [Google Scholar] [CrossRef]
- Pastorek, L.; Venit, T.; Hozak, P. Holography microscopy as an artifact-free alternative to phase-contrast. Histochem. Cell Biol. 2018, 149, 179–186. [Google Scholar] [CrossRef]
- Kasprowicz, R.; Suman, R.; O’Toole, P. Characterising live cell behaviour: Traditional label-free and quantitative phase imaging approaches. Int. J. Biochem. Cell Biol. 2017, 84, 89–95. [Google Scholar] [CrossRef] [PubMed]
- El-Schich, Z.; Molder, A.; Tassidis, H.; Harkonen, P.; Miniotis, M.F.; Wingren, A.G. Induction of morphological changes in death-induced cancer cells monitored by holographic microscopy. J. Struct. Biol. 2015, 189, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Balvan, J.; Krizova, A.; Gumulec, J.; Raudenska, M.; Sladek, Z.; Sedlackova, M. Multimodal Holographic Microscopy: Distinction between Apoptosis and Oncosis (vol 10, e0121674, 2015). PLoS ONE 2015, 10, e0121674. [Google Scholar] [CrossRef] [Green Version]
- Vicar, T.; Raudenska, M.; Gumulec, J.; Masarik, M.; Balvan, J. Detection and characterization of apoptotic and necrotic cell death by time-lapse quantitative phase image analysis. bioRxiv 2019.
- Barker, K.L.; Boucher, K.M.; Judson-Torres, R.L. Label-free classification of apoptosis, ferroptosis and necroptosis using digital holographic cytometry. Appl. Sci. 2020, 10, 4439. [Google Scholar] [CrossRef]
- Fojtu, M.; Balvan, J.; Raudenska, M.; Vicar, T.; Bousa, D.; Sofer, Z.; Masarik, M.; Pumera, M. Black Phosphorus Cytotoxicity Assessments Pitfalls: Advantages and Disadvantages of Metabolic and Morphological Assays. Chem. Eur. J. 2019, 25, 349–360. [Google Scholar] [CrossRef]
- Sternbæk, L.; Kimani, M.; Gawlitza, K.; Janicke, B.; Alm, K.; Wingren, A. Digital Holographic Cytometry: Macrophage Uptake of Nanoprobes. Imaging Microsc. 2019, 1, 21–23. [Google Scholar]
- Wingren, A.G. Moving into a new dimension: Tracking migrating cells with digital holographic cytometry in 3D. Cytom. Part A 2018, 95, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Feith, M.; Janicke, B.; Alm, K.; El-Schich, Z. Evaluation of the impact of imprinted polymer particles on morphology and motility of breast cancer cells by using digital holographic cytometry. Appl. Sci. 2020, 10, 750. [Google Scholar] [CrossRef] [Green Version]
- Tolde, O.; Gandalovičová, A.; Křížová, A.; Veselý, P.; Chmelík, R.; Rosel, D.; Brábek, J. Quantitative phase imaging unravels new insight into dynamics of mesenchymal and amoeboid cancer cell invasion. Sci. Rep. 2018, 8, 12020. [Google Scholar] [CrossRef] [Green Version]
- El-Schich, Z.; Janicke, B.; Alm, K.; Dizeyi, N.; Persson, J.; Wingren, A. Discrimination between Breast Cancer Cells and White Blood Cells by Non-Invasive Measurements: Implications for a Novel In Vitro-Based Circulating Tumor Cell Model Using Digital Holographic Cytometry. Appl. Sci. 2020, 10, 4854. [Google Scholar] [CrossRef]
- Yamada, T.; Matsuda, A.; Koizumi, M.; Shinji, S.; Takahashi, G.; Iwai, T.; Takeda, K.; Ueda, K.; Yokoyama, Y.; Hara, K.; et al. Liquid Biopsy for the Management of Patients with Colorectal Cancer. Digestion 2019, 99, 39–45. [Google Scholar] [CrossRef]
- Ranc, V.; Srovnal, J.; Kvitek, L.; Hajduch, M. Discrimination of circulating tumor cells of breast cancer and colorectal cancer from normal human mononuclear cells using Raman spectroscopy. Analyst 2013, 138, 5983–5988. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Chen, P.; Wu, C.H.; Hoshino, K.; Sokolov, K.; Lane, N.; Liu, H.Y.; Huebschman, M.; Frenkel, E.; Zhang, J.X.J. Screening and Molecular Analysis of Single Circulating Tumor Cells Using Micromagnet Array. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef]
- Zhang, H.; Fu, X.; Hu, J.Y.; Zhu, Z.J. Microfluidic bead-based multienzyme-nanoparticle amplification for detection of circulating tumor cells in the blood using quantum dots labels. Anal. Chim. Acta 2013, 779, 64–71. [Google Scholar] [CrossRef]
- Gazouli, M.; Lyberopoulou, A.; Pericleous, P.; Rizos, S.; Aravantinos, G.; Nikiteas, N.; Anagnou, N.P.; Efstathopoulos, E.P. Development of a quantum-dot-labelled magnetic immunoassay method for circulating colorectal cancer cell detection. World J. Gastroenterol. 2012, 18, 4419–4426. [Google Scholar] [CrossRef]
- Vaiopoulos, A.G.; Kostakis, I.D.; Gkioka, E.; Athanasoula, K.C.; Pikoulis, E.; Papalambros, A.; Christopoulos, P.; Gogas, H.; Kouraklis, G.; Koutsilieris, M. Detection of Circulating Tumor Cells in Colorectal and Gastric Cancer Using a Multiplex PCR Assay. Anticancer Res. 2014, 34, 3083–3092. [Google Scholar] [PubMed]
- Merola, F.; Memmolo, P.; Miccio, L.; Mugnano, M.; Ferraro, P. Phase contrast tomography at lab on chip scale by digital holography. Methods 2018, 136, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Balasubramani, V.; Kujawińska, M.; Allier, C.; Anand, V.; Cheng, C.-J.; Depeursinge, C.; Hai, N.; Juodkazis, S.; Kalkman, J.; Kuś, A. Roadmap on digital holography-based quantitative phase imaging. J. Imaging 2021, 7, 252. [Google Scholar] [CrossRef] [PubMed]
- Balasubramani, V.; Kuś, A.; Tu, H.-Y.; Cheng, C.-J.; Baczewska, M.; Krauze, W.; Kujawińska, M. Holographic tomography: Techniques and biomedical applications. Appl. Opt. 2021, 60, B65–B80. [Google Scholar] [CrossRef]
- Merola, F.; Memmolo, P.; Miccio, L.; Savoia, R.; Mugnano, M.; Fontana, A.; D’ippolito, G.; Sardo, A.; Iolascon, A.; Gambale, A. Tomographic flow cytometry by digital holography. Light Sci. Appl. 2017, 6, e16241. [Google Scholar] [CrossRef] [Green Version]
- Nissim, N.; Dudaie, M.; Barnea, I.; Shaked, N.T. Real-Time Stain-Free Classification of Cancer Cells and Blood Cells Using Interferometric Phase Microscopy and Machine Learning. Cytom. Part A 2021, 99, 511–523. [Google Scholar] [CrossRef]
- Habaza, M.; Kirschbaum, M.; Guernth-Marschner, C.; Dardikman, G.; Barnea, I.; Korenstein, R.; Duschl, C.; Shaked, N.T. Rapid 3D refractive-index imaging of live cells in suspension without labeling using dielectrophoretic cell rotation. Adv. Sci. 2017, 4, 1600205. [Google Scholar] [CrossRef]
- Dudaie, M.; Nissim, N.; Barnea, I.; Gerling, T.; Duschl, C.; Kirschbaum, M.; Shaked, N.T. Label-free discrimination and selection of cancer cells from blood during flow using holography-induced dielectrophoresis. J. Biophotonics 2020, 13, e202000151. [Google Scholar] [CrossRef]
- Ben Baruch, S.; Rotman-Nativ, N.; Baram, A.; Greenspan, H.; Shaked, N.T. Cancer-Cell Deep-Learning Classification by Integrating Quantitative-Phase Spatial and Temporal Fluctuations. Cells 2021, 10, 3353. [Google Scholar] [CrossRef]
- Allen, J.E.; El-Deiry, W.S. Circulating Tumor Cells and Colorectal Cancer. Curr. Color. Cancer Rep. 2010, 6, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Nicolazzo, C.; Massimi, I.; Lotti, L.V.; Vespa, S.; Raimondi, C.; Pulcinelli, F.M.; Gradilone, A.; Gazzaniga, P. Impact of chronic exposure to bevacizumab on EpCAM-based detection of circulating tumor cells. Chin. J. Cancer Res. 2015, 27, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Ravindran, B. Isolation of Human PBMCs. Bio-Protoc. 2013, 3, e323. [Google Scholar] [CrossRef]
- El-Schich, Z.; Leida Mölder, A.; Gjörloff Wingren, A. Quantitative phase imaging for label-free analysis of cancer cells—Focus on digital holographic microscopy. Appl. Sci. 2018, 8, 1027. [Google Scholar] [CrossRef] [Green Version]
- El-Schich, Z.; Zhang, Y.; Göransson, T.; Dizeyi, N.; Persson, J.; Johansson, E.; Caraballo, R.; Elofsson, M.; Shinde, S.; Sellergren, B.; et al. Sialic Acid as a Biomarker Studied in Breast Cancer Cell Lines In Vitro Using Fluorescent Molecularly Imprinted Polymers. Appl. Sci. 2021, 11, 3256. [Google Scholar] [CrossRef]
- Levesque, M.C.; Haynes, B.F. In vitro culture of human peripheral blood monocytes induces hyaluronan binding and up-regulates monocyte variant CD44 isoform expression. J. Immunol. 1996, 156, 1557. [Google Scholar]
- Keller, L.; Werner, S.; Pantel, K. Biology and clinical relevance of EpCAM. Cell Stress 2019, 3, 165. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.A.; Mitchell, E.; Miller, M.C.; et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 2009, 20, 1223–1229. [Google Scholar] [CrossRef]
- Singh, D.K.; Ahrens, C.C.; Li, W.; Vanapalli, S.A. Label-free, high-throughput holographic screening and enumeration of tumor cells in blood. Lab Chip 2017, 17, 2920–2932. [Google Scholar] [CrossRef]
- Park, J.Y.; Jeong, A.L.; Joo, H.J.; Han, S.; Kim, S.-H.; Kim, H.-Y.; Lim, J.-S.; Lee, M.-S.; Choi, H.-K.; Yang, Y. Development of suspension cell culture model to mimic circulating tumor cells. Oncotarget 2017, 9, 622–640. [Google Scholar] [CrossRef] [Green Version]
- Lam, V.K.; Nguyen, T.C.; Chung, B.M.; Nehmetallah, G.; Raub, C.B. Quantitative assessment of cancer cell morphology and motility using telecentric digital holographic microscopy and machine learning. Cytom. Part A 2018, 93, 334–345. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Politaki, E.; Agelaki, S.; Apostolaki, S.; Hatzidaki, D.; Strati, A.; Koinis, F.; Perraki, M.; Saloustrou, G.; Stoupis, G.; Kallergi, G.; et al. A Comparison of Three Methods for the Detection of Circulating Tumor Cells in Patients with Early and Metastatic Breast Cancer. Cell. Physiol. Biochem. 2017, 44, 594–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bocanegra Evans, H.; Gorumlu, S.; Aksak, B.; Castillo, L.; Sheng, J. Holographic microscopy and microfluidics platform for measuring wall stress and 3D flow over surfaces textured by micro-pillars. Sci. Rep. 2016, 6, 28753. [Google Scholar] [CrossRef]
- Armistead, F.J.; Gala De Pablo, J.; Gadêlha, H.; Peyman, S.A.; Evans, S.D. Cells Under Stress: An Inertial-Shear Microfluidic Determination of Cell Behavior. Biophys. J. 2019, 116, 1127–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Mahjoubfar, A.; Tai, L.-C.; Blaby, I.; Huang, A.; Niazi, K.; Jalali, B. Deep Learning in Label-free Cell Classification. Sci. Rep. 2016, 6, 21471. [Google Scholar] [CrossRef]



| Pure Suspension PBMC | Pure Suspension COLO 205 | Mixed Suspension PBMC (90%) and COLO 205 (10%) | Mixed Suspension PBMC (95%) and COLO 205 (5%) | |||
|---|---|---|---|---|---|---|
| Gated cells | 2488 | 2436 | 1512 | 185 | 1736 | 104 |
| % gated cells | 100 | 100 | 89 | 11 | 94 | 6 |
| Area (µm2) | 36.5 | 162.7 | 33.6 | 163.5 | 38.2 | 169.3 |
| Optical thickness (µm) | 4.1 | 8.8 | 4.6 | 8.3 | 4.1 | 8.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feith, M.; Zhang, Y.; Persson, J.L.; Balvan, J.; El-Schich, Z.; Wingren, A.G. Circulating Tumor Cell Models Mimicking Metastasizing Cells In Vitro: Discrimination of Colorectal Cancer Cells and White Blood Cells Using Digital Holographic Cytometry. Photonics 2022, 9, 955. https://doi.org/10.3390/photonics9120955
Feith M, Zhang Y, Persson JL, Balvan J, El-Schich Z, Wingren AG. Circulating Tumor Cell Models Mimicking Metastasizing Cells In Vitro: Discrimination of Colorectal Cancer Cells and White Blood Cells Using Digital Holographic Cytometry. Photonics. 2022; 9(12):955. https://doi.org/10.3390/photonics9120955
Chicago/Turabian StyleFeith, Marek, Yuecheng Zhang, Jenny L. Persson, Jan Balvan, Zahra El-Schich, and Anette Gjörloff Wingren. 2022. "Circulating Tumor Cell Models Mimicking Metastasizing Cells In Vitro: Discrimination of Colorectal Cancer Cells and White Blood Cells Using Digital Holographic Cytometry" Photonics 9, no. 12: 955. https://doi.org/10.3390/photonics9120955
APA StyleFeith, M., Zhang, Y., Persson, J. L., Balvan, J., El-Schich, Z., & Wingren, A. G. (2022). Circulating Tumor Cell Models Mimicking Metastasizing Cells In Vitro: Discrimination of Colorectal Cancer Cells and White Blood Cells Using Digital Holographic Cytometry. Photonics, 9(12), 955. https://doi.org/10.3390/photonics9120955