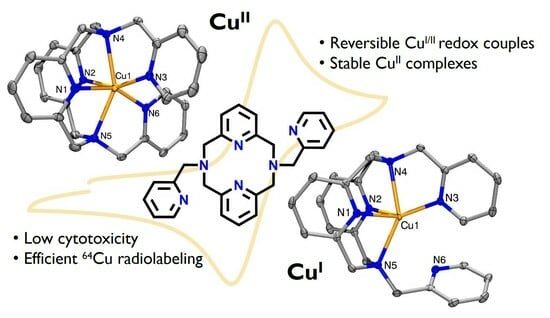
Pentadentate and Hexadentate Pyridinophane Ligands Support Reversible Cu(II)/Cu(I) Redox Couples
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis
2.2. Structural Characterization of Metal Complexes
2.3. Complex Characterization
2.4. Ligand Acidity Constants and Complex Stability Constants
2.5. Radiolabeling Studies
2.6. Cytotoxicity Studies
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krylova, K.; Kulatilleke, C.P.; Heeg, M.J.; Salhi, C.A.; Ochrymowycz, L.A.; Rorabacher, D.B. A Structural Strategy for Generating Rapid Electron-Transfer Kinetics in Copper(II/I) Systems. Inorg. Chem. 1999, 38, 4322. [Google Scholar] [CrossRef]
- Rorabacher, D.B. Electron Transfer by Copper Centers. Chem. Rev. 2004, 104, 651. [Google Scholar] [CrossRef]
- Himes, R.A.; Karlin, K.D. Copper–dioxygen complex mediated C–H bond oxygenation: Relevance for particulate methane monooxygenase (pMMO). Curr. Opin. Chem. Biol. 2009, 13, 119. [Google Scholar] [CrossRef] [PubMed]
- Mirica, L.M.; Ottenwaelder, X.; Stack, T.D.P. Structure and Spectroscopy of Copper-Dioxygen Complexes. Chem. Rev. 2004, 104, 1013. [Google Scholar] [CrossRef]
- You, Y.; Han, Y.; Lee, Y.-M.; Park, S.Y.; Nam, W.; Lippard, S.J. Phosphorescent Sensor for Robust Quantification of Copper(II) Ion. J. Am. Chem. Soc. 2011, 133, 11488. [Google Scholar] [CrossRef]
- Jung, H.S.; Kwon, P.S.; Lee, J.W.; Kim, J.I.; Hong, C.S.; Kim, J.W.; Yan, S.; Lee, J.Y.; Lee, J.H.; Joo, T.; et al. Coumarin-Derived Cu2+-Selective Fluorescence Sensor: Synthesis, Mechanisms, and Applications in Living Cells. J. Am. Chem. Soc. 2009, 131, 2008. [Google Scholar] [CrossRef] [PubMed]
- Terpstra, K.; Wang, Y.; Huynh, T.T.; Bandara, N.; Cho, H.-J.; Rogers, B.E.; Mirica, L.M. Divalent 2-(4-Hydroxyphenyl)benzothiazole Bifunctional Chelators for 64Cu PET Imaging in Alzheimer’s Disease. Inorg. Chem. 2022, 61, 20326–20336. [Google Scholar] [CrossRef]
- Cho, H.J.; Huynh, T.T.; Rogers, B.E.; Mirica, L.M. Design of a multivalent bifunctional chelator for diagnostic (64)Cu PET imaging in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2020, 117, 30928. [Google Scholar] [CrossRef] [PubMed]
- Guillou, A.; Lima, L.M.P.; Esteban-Gómez, D.; Le Poul, N.; Bartholomä, M.D.; Platas-Iglesias, C.; Delgado, R.; Patinec, V.; Tripier, R. Methylthiazolyl Tacn Ligands for Copper Complexation and Their Bifunctional Chelating Agent Derivatives for Bioconjugation and Copper-64 Radiolabeling: An Example with Bombesin. Inorg. Chem. 2019, 58, 2669. [Google Scholar] [CrossRef]
- Pena-Bonhome, C.; Fiaccabrino, D.; Rama, T.; Fernández-Pavón, D.; Southcott, L.; Zhang, Z.; Lin, K.-S.; de Blas, A.; Patrick, B.O.; Schaffer, P.; et al. Toward 68Ga and 64Cu Positron Emission Tomography Probes: Is H2dedpa-N,N′-pram the Missing Link for dedpa Conjugation? Inorg. Chem. 2023; ASAP. [Google Scholar] [CrossRef]
- Cai, Z.; Anderson, C.J. Chelators for copper radionuclides in positron emission tomography radiopharmaceuticals. J. Label. Compd. Radiopharm. 2014, 57, 224. [Google Scholar] [CrossRef]
- Bandara, N.; Sharma, A.K.; Krieger, S.; Schultz, J.W.; Han, B.H.; Rogers, B.E.; Mirica, L.M. Evaluation of 64Cu-based Radiopharmaceuticals That Target Aβ Peptide Aggregates as Diagnostic Tools for Alzheimer’s Disease. J. Am. Chem. Soc. 2017, 139, 12550. [Google Scholar] [CrossRef] [PubMed]
- Morfin, J.-F.; Lacerda, S.; Geraldes, C.F.G.C.; Tóth, É. Metal complexes for the visualisation of amyloid peptides. Sens. Diagn. 2022, 1, 627. [Google Scholar] [CrossRef]
- Uzal-Varela, R.; Patinec, V.; Tripier, R.; Valencia, L.; Maneiro, M.; Canle, M.; Platas-Iglesias, C.; Esteban-Gómez, D.; Iglesias, E. On the dissociation pathways of copper complexes relevant as PET imaging agents. J. Inorg. Biochem. 2022, 236, 111951. [Google Scholar] [CrossRef]
- Tosato, M.; Franchi, S.; Isse, A.A.; Del Vecchio, A.; Zanoni, G.; Alker, A.; Asti, M.; Gyr, T.; Di Marco, V.; Mäcke, H. Is Smaller Better? Cu2+/Cu+ Coordination Chemistry and Copper-64 Radiochemical Investigation of a 1,4,7-Triazacyclononane-Based Sulfur-Rich Chelator. Inorg. Chem. 2023; ASAP. [Google Scholar] [CrossRef]
- Tosato, M.; Dalla Tiezza, M.; May, N.V.; Isse, A.A.; Nardella, S.; Orian, L.; Verona, M.; Vaccarin, C.; Alker, A.; Mäcke, H.; et al. Copper Coordination Chemistry of Sulfur Pendant Cyclen Derivatives: An Attempt to Hinder the Reductive-Induced Demetalation in 64/67Cu Radiopharmaceuticals. Inorg. Chem. 2021, 60, 11530. [Google Scholar] [CrossRef]
- Matz, D.L.; Jones, D.G.; Roewe, K.D.; Gorbet, M.J.; Zhang, Z.; Chen, Z.Q.; Prior, T.J.; Archibald, S.J.; Yin, G.C.; Hubin, T.J. Synthesis, structural studies, kinetic stability, and oxidation catalysis of the late first row transition metal complexes of 4,10-dimethyl-1,4,7,10-tetraazabicyclo[6.5.2]pentadecane. Dalton Trans. 2015, 44, 12210. [Google Scholar] [CrossRef] [PubMed]
- Knighton, R.C.; Troadec, T.; Mazan, V.; Le Saëc, P.; Marionneau-Lambot, S.; Le Bihan, T.; Saffon-Merceron, N.; Le Bris, N.; Chérel, M.; Faivre-Chauvet, A.; et al. Cyclam-Based Chelators Bearing Phosphonated Pyridine Pendants for 64Cu-PET Imaging: Synthesis, Physicochemical Studies, Radiolabeling, and Bioimaging. Inorg. Chem. 2021, 60, 2634. [Google Scholar] [CrossRef]
- Lima, L.M.P.; Halime, Z.; Marion, R.; Camus, N.; Delgado, R.; Platas-Iglesias, C.; Tripier, R. Monopicolinate Cross-Bridged Cyclam Combining Very Fast Complexation with Very High Stability and Inertness of Its Copper(II) Complex. Inorg. Chem. 2014, 53, 5269. [Google Scholar] [CrossRef]
- Hierlmeier, I.; Guillou, A.; Earley, D.F.; Linden, A.; Holland, J.P.; Bartholomä, M.D. HNODThia: A Promising Chelator for the Development of 64Cu Radiopharmaceuticals. Inorg. Chem. 2023; ASAP. [Google Scholar] [CrossRef]
- Brudenell, S.J.; Spiccia, L.; Tiekink, E.R.T. Binuclear Copper(II) Complexes of Bis(pentadentate) Ligands Derived from Alkyl-Bridged Bis(1,4,7-triazacyclonane) Macrocycles. Inorg. Chem. 1996, 35, 1974. [Google Scholar] [CrossRef]
- Khusnutdinova, J.R.; Luo, J.; Rath, N.P.; Mirica, L.M. Late First-Row Transition Metal Complexes of a Tetradentate Pyridinophane Ligand: Electronic Properties and Reactivity Implications. Inorg. Chem. 2013, 52, 3920. [Google Scholar] [CrossRef] [PubMed]
- Wessel, A.J.; Schultz, J.W.; Tang, F.; Duan, H.; Mirica, L.M. Improved synthesis of symmetrically & asymmetrically N-substituted pyridinophane derivatives. Org. Biomol. Chem. 2017, 15, 9923. [Google Scholar]
- Huang, Y.; Huynh, T.T.; Sun, L.; Hu, C.-H.; Wang, Y.-C.; Rogers, B.E.; Mirica, L.M. Neutral Ligands as Potential 64Cu Chelators for Positron Emission Tomography Imaging Applications in Alzheimer’s Disease. Inorg. Chem. 2022, 61, 4778. [Google Scholar] [CrossRef] [PubMed]
- Halfen, J.A.; Tolman, W.B.; Weighardt, K. C2-Symmetric 1,4-Diisopropyl-7-R-1,4,7-Triazacyclononanes. In Inorganic Syntheses; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2007. [Google Scholar]
- Li, Y.; Lu, X.-M.; Sheng, X.; Lu, G.-Y.; Shao, Y.; Xu, Q. DNA cleavage promoted by Cu2+ complex of cyclen containing pyridine subunit. J. Inclusion Phenom. Macrocyclic Chem. 2007, 59, 91. [Google Scholar] [CrossRef]
- Che, C.M.; Li, Z.Y.; Wong, K.Y.; Poon, C.K.; Mak, T.C.W.; Peng, S.M. A simple synthetic route to N,N’-dialkyl-2,11-diaza[3.3](2,6)pyridinophanes. Crystal structures of N,N’-di-tert-butyl-2,11-diaza[3.3](2,6)pyridinophane and its copper(II) complex. Polyhedron 1994, 13, 771. [Google Scholar] [CrossRef]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen-sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2[prime or minute]-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 7, 1349. [Google Scholar] [CrossRef]
- Cotton, F.A.; Wilkinson, G. Advanced Inorganic Chemistry, 5th ed.; Wiley–Interscience: New York, NY, USA, 1988. [Google Scholar]
- Pratt, R.C.; Mirica, L.M.; Stack, T.D.P. Snapshots of a metamorphosing Cu(II) ground state in a galactose oxidase-inspired complex. Inorg. Chem. 2004, 43, 8030. [Google Scholar] [CrossRef]
- Bottino, F.; Di Grazia, M.; Finocchiaro, P.; Fronczek, F.R.; Mamo, A.; Pappalardo, S. Reaction of Tosylamide Monosodium Salt with Bis(halomethyl) Compounds: An Easy Entry to Symmetrical N-tosylazamacrocycles. J. Org. Chem. 1988, 53, 3521. [Google Scholar] [CrossRef]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad simulation and speciation (HySS): A utility program for the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311. [Google Scholar] [CrossRef]
- Green, K.N.; Pota, K.; Tircsó, G.; Gogolák, R.A.; Kinsinger, O.; Davda, C.; Blain, K.; Brewer, S.M.; Gonzalez, P.; Johnston, H.M.; et al. Dialing in on pharmacological features for a therapeutic antioxidant small molecule. Dalton Trans. 2019, 48, 12430. [Google Scholar] [CrossRef]
- Marlin, A.; Koller, A.; Madarasi, E.; Cordier, M.; Esteban-Gómez, D.; Platas-Iglesias, C.; Tircsó, G.; Boros, E.; Patinec, V.; Tripier, R. H3nota Derivatives Possessing Picolyl and Picolinate Pendants for Ga3+ Coordination and 67Ga3+ Radiolabeling. Inorg. Chem. 2023; ASAP. [Google Scholar] [CrossRef] [PubMed]
- Nonat, A.M.; Gateau, C.; Fries, P.H.; Helm, L.; Mazzanti, M. New Bisaqua Picolinate-Based Gadolinium Complexes as MRI Contrast Agents with Substantial High-Field Relaxivities. Eur. J. Inorg. Chem. 2012, 2012, 2049. [Google Scholar] [CrossRef]
- Nonat, A.; Gateau, C.; Fries, P.H.; Mazzanti, M. Lanthanide Complexes of a Picolinate Ligand Derived from 1,4,7-Triazacyclononane with Potential Application in Magnetic Resonance Imaging and Time-Resolved Luminescence Imaging. Chem. Eur. J. 2006, 12, 7133. [Google Scholar] [CrossRef]
- Brasse, D.; Nonat, A. Radiometals: Towards a new success story in nuclear imaging? Dalton Trans. 2015, 44, 4845. [Google Scholar] [CrossRef]
- Price, E.W.; Orvig, C. Matching chelators to radiometals for radiopharmaceuticals. Chem. Soc. Rev. 2014, 43, 260. [Google Scholar] [CrossRef]
- Wadas, T.J.; Wong, E.H.; Weisman, G.R.; Anderson, C.J. Coordinating Radiometals of Copper, Gallium, Indium, Yttrium, and Zirconium for PET and SPECT Imaging of Disease. Chem. Rev. 2010, 110, 2858. [Google Scholar] [CrossRef] [PubMed]
- Woodin, K.S.; Heroux, K.J.; Boswell, C.A.; Wong, E.H.; Weisman, G.R.; Niu, W.; Tomellini, S.A.; Anderson, C.J.; Zakharov, L.N.; Rheingold, A.L. Kinetic Inertness and Electrochemical Behavior of Copper(II) Tetraazamacrocyclic Complexes: Possible Implications for in Vivo Stability. Eur. J. Inorg. Chem. 2005, 2005, 4829. [Google Scholar] [CrossRef]
- Anderegg, G.; Arnaud-Neu, F.; Delgado, R.; Felcman, J.; Popov, K. Critical evaluation of stability constants of metal complexes of complexones for biomedical and environmental applications* (IUPAC Technical Report). Pure Appl. Chem. 2005, 77, 1445. [Google Scholar] [CrossRef]
- Martell, A.E.; Motekaitis, R.J.; Clarke, E.T.; Delgado, R.; Sun, Y.; Ma, R. Stability constants of metal complexes of macrocyclic ligands with pendant donor groups. Supramol. Chem. 1996, 6, 353. [Google Scholar] [CrossRef]
- Irangu, J.; Ferguson, M.J.; Jordan, R.B. Reaction of copper(II) with ferrocene and 1,1 ‘-dimethylferrocene in aqueous acetonitrile: The copper(II/I) self-exchange rate. Inorg. Chem. 2005, 44, 1619. [Google Scholar] [CrossRef]
- Evans, D.F. Determination of the Paramagnetic Susceptibility of Substances in Solution By NMR. J. Chem. Soc. 1959, 2003. [Google Scholar] [CrossRef]
- De Buysser, K.; Herman, G.G.; Bruneel, E.; Hoste, S.; Van Driessche, I. Determination of the Number of Unpaired Electrons in Metal-Complexes. A Comparison Between the Evans’ Method and Susceptometer Results. Chem. Phys. 2005, 315, 286. [Google Scholar] [CrossRef]
- Bain, G.A.; Berry, J.F. Diamagnetic Corrections and Pascal’s Constants. J. Chem. Educ. 2008, 85, 532. [Google Scholar] [CrossRef]
- Krause, L.; Herbst-Irmer, R.; Sheldrick, G.M.; Stalke, D. Comparison of silver and molybdenum microfocus X-ray sources for single-crystal structure determination. J. Appl. Crystallogr. 2015, 48, 3. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. Sect. A 2015, 71, 3. [Google Scholar] [CrossRef]
- Bagchi, P.; Morgan, M.T.; Bacsa, J.; Fahrni, C.J. Robust Affinity Standards for Cu(I) Biochemistry. J. Am. Chem. Soc. 2013, 135, 18549. [Google Scholar] [CrossRef]









| 12+ | 2+ | 32+ | 4+ | |
|---|---|---|---|---|
| Cu-N1 | 2.056(4) | 2.1341(1) | 1.944(9) | 2.1286(1) |
| Cu-N2 | 2.028(4) | 2.0817(1) | 2.173(7) | 2.0768(2) |
| Cu-N3 | 2.003(4) | 1.9640(1) | 1.967(8) | 1.9461(1) |
| Cu-N4 | 2.276(4) | 2.3983(1) | 2.258(7) | 2.3957(2) |
| Cu-N5 | 2.348(4) | 2.3456(1) | 2.165(8) | 2.262(2) |
| Cu-N6 | 2.017(4) | 3.323 | 2.219(9) | --- |
| N2-Cu-N1 | 84.43 | 81.36 | 82.9 | 83.07 |
| N4-Cu-N5 | 148.38 | 146.78 | 152.1 | 147.85 |
| φ(°) a | 86.60, 84.19 | 86.91, 87.93 | 88.07, 89.28 | 88.19, 87.00 |
| θ(°) b | 36.19 | 28.92 | 20.39 | 31.80 |
| gx | gy | gz | Az (G) | |
|---|---|---|---|---|
| 1·(OTf)2 | 2.070 | 2.055 | 2.259 | 145 |
| 3·(OTf)2 | 2.067 | 2.056 | 2.264 | 152 |
| 12+ | 2+ | 32+ | 4+ |
|---|---|---|---|
| E, V (ΔEp, mV) a,b,c | |||
| E1/2 = −0.752 (97) Eox = 1.047 | E1/2 = −0.716 (176) | E1/2 = −0.468 (105) Eox = 1.552 | E1/2 = −0.441 (96) |
| UV-Vis, λmax, nm (ε, M−1 cm−1), MeCN | |||
| 257 (22,775), 340 (446), 717 (146) | 250 (8395), 362 (2438), 444 (893) | 258 (12,251), 322 (671), 687 (91) | 246 (11,731), 332 (3331), 370 (3854), 435 (1611) |
| μeff (μB) at 293 K, Evans’ Method, CD3CN | |||
| 1.80 | N/A | 1.71 | N/A |
| PicN4 | PicMeN4 | |
|---|---|---|
| [H4L]4+ = [H3L]3+ + H+ | - | 2.47(9) |
| [H3L]3+ = [H2L]2+ + H+ | 3.60(3) | 5.46(9) |
| [H2L]2+ = [HL]+ + H+ | 5.32(0) | 9.16(9) |
| [HL] = [L] + H+ | 8.94(6) | 11.13(8) |
| PicN4 + Cu2+ | PicMeN4 + Cu2+ | PicN4 + Zn2+ | PicMeN4 + Zn2+ | |
|---|---|---|---|---|
| M2+ + H2L+ = [MH2L]4+ | 4.13(3) | - | - | - |
| M2+ + HL+ = [MHL]3+ | 7.40(1) | 4.54(1) | 2.67(2) | 9.28(7) |
| M2+ + L = [ML]2+ | 17.96(3) | 17.07(1) | 11.45(4) | 10.41(7) |
| [ML(H2O)]2+ = [ML(OH)]+ + H+ | - | 7.75(2) | - | - |
| pM2+ (pH 7.4)a | 16.81 | 12.90 | 8.87 | 8.02 |
| log(KCu(II)L) | 17.96 | 17.07 | - | - |
| log(KCu(I)L) | 7.05 | 9.46 | - | - |
| Chelator | Log(KCu(II)L) | Ref. |
|---|---|---|
| PicN4 | 17.96 | This work |
| PicMeN4 | 17.07 | This work |
| YW-15-Me | 14.7 | [7] |
| DO4S | 19.6 | [16] |
| PCTA | 19.1 | [37,38] |
| EDTA | 19.2 | [37,39,40] |
| TETA | 21.1 | [39,40,41,42] |
| DOTA | 22.2 | [39,40,41] |
| cyclen | 24.6 | [39,40] |
| 64Cu Complex | MW (g/mol) | log Doct |
|---|---|---|
| 64Cu-PicN4 | 556.98 | −1.564 ± 0.26 |
| 64Cu-PicMeN4 | 479.90 | −2.171 ± 0.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blade, G.; Wessel, A.J.; Terpstra, K.; Mirica, L.M. Pentadentate and Hexadentate Pyridinophane Ligands Support Reversible Cu(II)/Cu(I) Redox Couples. Inorganics 2023, 11, 446. https://doi.org/10.3390/inorganics11110446
Blade G, Wessel AJ, Terpstra K, Mirica LM. Pentadentate and Hexadentate Pyridinophane Ligands Support Reversible Cu(II)/Cu(I) Redox Couples. Inorganics. 2023; 11(11):446. https://doi.org/10.3390/inorganics11110446
Chicago/Turabian StyleBlade, Glenn, Andrew J. Wessel, Karna Terpstra, and Liviu M. Mirica. 2023. "Pentadentate and Hexadentate Pyridinophane Ligands Support Reversible Cu(II)/Cu(I) Redox Couples" Inorganics 11, no. 11: 446. https://doi.org/10.3390/inorganics11110446
APA StyleBlade, G., Wessel, A. J., Terpstra, K., & Mirica, L. M. (2023). Pentadentate and Hexadentate Pyridinophane Ligands Support Reversible Cu(II)/Cu(I) Redox Couples. Inorganics, 11(11), 446. https://doi.org/10.3390/inorganics11110446