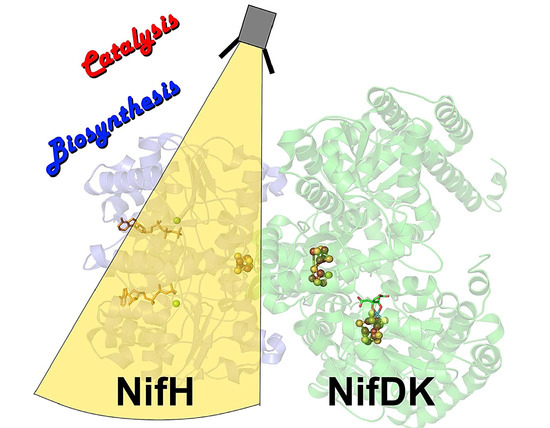
The Fe Protein: An Unsung Hero of Nitrogenase
Abstract
:1. Introduction
2. The Roles of the Fe Protein
2.1. Mo and Homocitrate Insertase
2.2. P-Cluster Formation
2.3. Electron Transfer for Nitrogenase Catalysis
2.4. Adventitious Reactivity of Fe Proteins
3. Features of the Fe Protein
3.1. Nucleotide Binding to NifH
3.2. The NifH and NifDK Protein Complex
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Burgess, B.K.; Lowe, D.J. Mechanism of molybdenum nitrogenase. Chem. Rev. 1996, 96, 2983–3012. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, M.; Komiya, H.; Chakrabarti, P.; Woo, D.; Kornuc, J.; Rees, D. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 1992, 257, 1653–1659. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.-J.; Peters, J.W.; Lanzilotta, W.N.; Ryle, M.J.; Seefeldt, L.C.; Howard, J.B.; Rees, D.C. MgAPT-bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127δ-Fe-protein and the MoFe-protein. Biochemistry 2001, 40, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Corbett, M.C.; Fay, A.W.; Webber, J.A.; Hodgson, K.O.; Hedman, B.; Ribbe, M.W. Nitrogenase Fe protein: A molybdate/homocitrate insertase. Proc. Natl. Acad. Sci. USA 2006, 103, 17125–17130. [Google Scholar] [CrossRef] [PubMed]
- Ribbe, M.W.; Hu, Y.; Guo, M.; Schmid, B.; Burgess, B.K. The femoco-deficient MoFe protein produced by a nifH deletion strain of Azotobacter vinelandii shows unusual P-cluster features. J. Biol. Chem. 2002, 277, 23469–23476. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Blank, M.A.; Fay, A.W.; Yoshizawa, J.M.; Hu, Y.; Hodgson, K.O.; Hedman, B.; Ribbe, M.W. Stepwise formation of P-cluster in nitrogenase MoFe protein. Proc. Natl. Acad. Sci. USA 2009, 106, 18474–18478. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.M.; Ludden, P.W. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 2008, 62, 93–111. [Google Scholar] [CrossRef] [PubMed]
- Eady, R.R. Structure–function relationships of alternative nitrogenases. Chem. Rev. 1996, 96, 3013–3030. [Google Scholar] [CrossRef] [PubMed]
- Ribbe, M.W.; Hu, Y.; Hodgson, K.O.; Hedman, B. Biosynthesis of nitrogenase metalloclusters. Chem. Rev. 2014, 114, 4063–4080. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fay, A.W.; Ribbe, M.W. Identification of a nitrogenase FeMo cofactor precursor on NifEN complex. Proc. Natl. Acad. Sci. USA 2005, 102, 3236–3241. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, J.T.; Hu, Y.; Wiig, J.A.; Rees, D.C.; Ribbe, M.W. Structure of precursor-bound NifEN: A nitrogenase femo cofactor maturase/insertase. Science 2011, 331, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Corbett, M.C.; Fay, A.W.; Webber, J.A.; Hodgson, K.O.; Hedman, B.; Ribbe, M.W. FeMo cofactor maturation on NifEN. Proc. Natl. Acad. Sci. USA 2006, 103, 17119–17124. [Google Scholar] [CrossRef] [PubMed]
- Fay, A.W.; Wiig, J.A.; Lee, C.C.; Hu, Y. Identification and characterization of functional homologs of nitrogenase cofactor biosynthesis protein NifB from methanogens. Proc. Natl. Acad. Sci. USA 2015, 112, 14829–14833. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.A.; Curatti, L.; Aznar, C.P.; Perova, Z.; Britt, R.D.; Rubio, L.M. Metal trafficking for nitrogen fixation: NifQ donates molybdenum to NifEN/NifH for the biosynthesis of the nitrogenase FeMo-cofactor. Proc. Natl. Acad. Sci. USA 2008, 105, 11679–11684. [Google Scholar] [CrossRef] [PubMed]
- Bjornsson, R.; Lima, F.A.; Spatzal, T.; Weyhermuller, T.; Glatzel, P.; Bill, E.; Einsle, O.; Neese, F.; DeBeer, S. Identification of a spin-coupled Mo(III) in the nitrogenase iron-molybdenum cofactor. Chem. Sci. 2014, 5, 3096–3103. [Google Scholar] [CrossRef]
- Fisher, K.; Lowe, D.J.; Petersen, J. Vanadium(v) is reduced by the ‘as isolated’ nitrogenase Fe-protein at neutral pH. Chem. Commun. 2006, 2807–2809. [Google Scholar] [CrossRef]
- Broach, R.B.; Rupnik, K.; Hu, Y.; Fay, A.W.; Cotton, M.; Ribbe, M.W.; Hales, B.J. Variable-temperature, variable-field magnetic circular dichroism spectroscopic study of the metal clusters in the ΔnifB and ΔnifH MoFe proteins of nitrogenase from Azotobacter vinelandii. Biochemistry 2006, 45, 15039–15048. [Google Scholar] [CrossRef] [PubMed]
- Corbett, M.C.; Hu, Y.; Naderi, F.; Ribbe, M.W.; Hedman, B.; Hodgson, K.O. Comparison of iron-molybdenum cofactor-deficient nitrogenase MoFe proteins by X-ray absorption spectroscopy: Implications for P-cluster biosynthesis. J. Biol. Chem. 2004, 279, 28276–28282. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Corbett, M.C.; Fay, A.W.; Webber, J.A.; Hedman, B.; Hodgson, K.O.; Ribbe, M.W. Nitrogenase reactivity with P-cluster variants. Proc. Natl. Acad. Sci. USA 2005, 102, 13825–13830. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fay, A.W.; Dos Santos, P.C.; Naderi, F.; Ribbe, M.W. Characterization of Azotobacter vinelandii nifZ deletion strains: Indication of stepwise MoFe protein assembly. J. Biol. Chem. 2004, 279, 54963–54971. [Google Scholar] [CrossRef] [PubMed]
- Cotton, M.S.; Rupnik, K.; Broach, R.B.; Hu, Y.; Fay, A.W.; Ribbe, M.W.; Hales, B.J. VTVH-MCD study of the ΔnifBΔnifZ MoFe protein from Azotobacter vinelandii. J. Am. Chem. Soc. 2009, 131, 4558–4559. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.A.; Harris, D.F.; Wilker, M.B.; Rasmussen, A.; Khadka, N.; Hamby, H.; Keable, S.; Dukovic, G.; Peters, J.W.; Seefeldt, L.C.; et al. Light-driven dinitrogen reduction catalyzed by a CdS:Nitrogenase MoFe protein biohybrid. Science 2016, 352, 448–450. [Google Scholar] [CrossRef] [PubMed]
- Tanifuji, K.; Lee, C.C.; Ohki, Y.; Tatsumi, K.; Hu, Y.; Ribbe, M.W. Combining a nitrogenase scaffold and a synthetic compound into an artificial enzyme. Angew. Chem. Int. Ed. 2015, 54, 14022–14025. [Google Scholar] [CrossRef] [PubMed]
- Milton, R.D.; Abdellaoui, S.; Khadka, N.; Dean, D.R.; Leech, D.; Seefeldt, L.C.; Minteer, S.D. Nitrogenase bioelectrocatalysis: Heterogeneous ammonia and hydrogen production by MoFe protein. Energy Environ. Sci. 2016, 9, 2550–2554. [Google Scholar] [CrossRef]
- Rebelein, J.G.; Hu, Y.; Ribbe, M.W. Widening the product profile of carbon dioxide reduction by vanadium nitrogenase. ChemBioChem 2015, 16, 1993–1996. [Google Scholar] [CrossRef] [PubMed]
- Yates, M.G. Electron transport to nitrogenase in Azotobacter chroococcum: Azotobacter flavodoxin hydroquinone as an electron donor. FEBS Lett. 1972, 27, 63–67. [Google Scholar] [CrossRef]
- Duyvis, M.G.; Wassink, H.; Haaker, H. Nitrogenase of Azotobacter vinelandii: Kinetic analysis of the Fe protein redox cycle. Biochemistry 1998, 37, 17345–17354. [Google Scholar] [CrossRef] [PubMed]
- Bennett, L.T.; Jacobson, M.R.; Dean, D.R. Isolation, sequencing, and mutagenesis of the nifF gene encoding flavodoxin from Azotobacter vinelandii. J. Biol. Chem. 1988, 263, 1364–1369. [Google Scholar] [PubMed]
- Thorneley, R.N.; Deistung, J. Electron-transfer studies involving flavodoxin and a natural redox partner, the iron protein of nitrogenase. Conformational constraints on protein-protein interactions and the kinetics of electron transfer within the protein complex. Biochem. J. 1988, 253, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.E.; Burgess, B.K.; Iismaa, S.E.; Smartt, C.T.; Jacobson, M.R.; Dean, D.R. Construction and characterization of an Azotobacter vinelandii strain with mutations in the genes encoding flavodoxin and ferredoxin I. J. Bacteriol. 1989, 171, 3162–3167. [Google Scholar] [CrossRef] [PubMed]
- Danyal, K.; Dean, D.R.; Hoffman, B.M.; Seefeldt, L.C. Electron transfer within nitrogenase: Evidence for a deficit-spending mechanism. Biochemistry 2011, 50, 9255–9263. [Google Scholar] [CrossRef] [PubMed]
- Watt, G.D.; Reddy, K.R.N. Formation of an all ferrous Fe4S4 cluster in the iron protein component of Azotobacter vinelandii nitrogenase. J. Inorg. Biochem. 1994, 53, 281–294. [Google Scholar] [CrossRef]
- Sickerman, N.S.; Hu, Y.; Ribbe, M.W. Activation of CO2 by vanadium nitrogenase. Chem. Asian J. 2017, 12, 1985–1996. [Google Scholar] [CrossRef] [PubMed]
- Rebelein, J.G.; Stiebritz, M.T.; Lee, C.C.; Hu, Y. Activation and reduction of carbon dioxide by nitrogenase iron proteins. Nat. Chem. Biol. 2016, 13, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, C.; Bali, A.; Blanco, G.; Contreras, A.; Drummond, M.; Merrick, M.; Walmsley, J.; Woodley, P. Regulation of expression of genes for three nitrogenases in Azotobacter vinelandii. In Nitrogen Fixation: Proceedings of the Fifth International Symposium on Nitrogen Fixation with Non-Legumes, Florence, Italy, 10–14 September 1990; Polsinelli, M., Materassi, R., Vincenzini, M., Eds.; Springer: Dordrecht, The Netherlands, 1991; pp. 13–23. [Google Scholar]
- Strop, P.; Takahara, P.M.; Chiu, H.-J.; Angove, H.C.; Burgess, B.K.; Rees, D.C. Crystal structure of the all-ferrous [4Fe-4S]0 form of the nitrogenase iron protein from Azotobacter vinelandii. Biochemistry 2001, 40, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, P.A.; Day, E.P.; Kent, T.A.; Orme-Johnson, W.H.; Münck, E. Mössbauer, epr, and magnetization studies of the Azotobacter vinelandii Fe protein. Evidence for a [4Fe-4S]1+ cluster with spin S = 3/2. J. Biol. Chem. 1985, 260, 11160–11173. [Google Scholar] [PubMed]
- Angove, H.C.; Yoo, S.J.; Burgess, B.K.; Münck, E. Mössbauer and EPR evidence for an all-ferrous Fe4S4 cluster with S = 4 in the Fe protein of nitrogenase. J. Am. Chem. Soc. 1997, 119, 8730–8731. [Google Scholar] [CrossRef]
- Yoo, S.J.; Angove, H.C.; Burgess, B.K.; Hendrich, M.P.; Münck, E. Mössbauer and integer-spin EPR studies and spin-coupling analysis of the [4Fe-4S]0 cluster of the Fe protein from Azotobacter vinelandii nitrogenase. J. Am. Chem. Soc. 1999, 121, 2534–2545. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Deng, L.; Holm, R.H.; Münck, E.; Bominaar, E.L. Mössbauer, electron paramagnetic resonance, and theoretical studies of a carbene-based all-ferrous Fe4S4 cluster: Electronic origin and structural identification of the unique spectroscopic site. Inorg. Chem. 2009, 48, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, M.; Münck, E.; Bominaar, E.L. Density functional theory study of an all ferrous 4Fe-4S cluster. Inorg. Chem. 2011, 50, 4322–4326. [Google Scholar] [CrossRef] [PubMed]
- Hiller, C.J.; Stiebritz, M.T.; Lee, C.C.; Liedtke, J.; Hu, Y. Tuning electron flux through nitrogenase with methanogen iron protein homologues. Chem. Eur. J. 2017, 23, 16152–16156. [Google Scholar] [CrossRef] [PubMed]
- Lowery, T.J.; Wilson, P.E.; Zhang, B.; Bunker, J.; Harrison, R.G.; Nyborg, A.C.; Thiriot, D.; Watt, G.D. Flavodoxin hydroquinone reduces Azotobacter vinelandii Fe protein to the all-ferrous redox state with a S = 0 spin state. Proc. Natl. Acad. Sci. USA 2006, 103, 17131–17136. [Google Scholar] [CrossRef] [PubMed]
- Onate, Y.A.; Finnegan, M.G.; Hales, B.J.; Johnson, M.K. Variable temperature magnetic circular dichroism studies of reduced nitrogenase iron proteins and [4Fe-4S]+ synthetic analog clusters. Biochim. Biophys. Acta Protein Struct. Mol. 1993, 1164, 113–123. [Google Scholar] [CrossRef]
- Blank, M.A.; Lee, C.C.; Hu, Y.; Hodgson, K.O.; Hedman, B.; Ribbe, M.W. Structural models of the [Fe4S4] clusters of homologous nitrogenase fe proteins. Inorg. Chem. 2011, 50, 7123–7128. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.B.; Seefeldt, L.C.; Peters, J.W. Insights into nucleotide signal transduction in nitrogenase: Structure of an iron protein with MgADP bound. Biochemistry 2000, 39, 14745–14752. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Krishnakumar, A.; McClead, J.; Johnson, M.K.; Seefeldt, L.C.; Szilagyi, R.K.; Peters, J.W. Insights into the role of nucleotide-dependent conformational change in nitrogenase catalysis: Structural characterization of the nitrogenase Fe protein Leu127 deletion variant with bound MgATP. J. Inorg. Biochem. 2006, 100, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Tezcan, F.A.; Kaiser, J.T.; Mustafi, D.; Walton, M.Y.; Howard, J.B.; Rees, D.C. Nitrogenase complexes: Multiple docking sites for a nucleotide switch protein. Science 2005, 309, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, H.; Kisker, C.; Schlessman, J.L.; Howard, J.B.; Rees, D.C. Structure of ADP·AlF4—Stabilized nitrogenase complex and its implications for signal transduction. Nature 1997, 387, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Robson, R.L. Identification of possible adenine nucleotide-binding sites in nitrogenase Fe- and MoFe-proteins by amino acid sequence comparison. FEBS Lett. 1984, 173, 394–398. [Google Scholar] [CrossRef]
- Walker, J.E.; Saraste, M.; Runswick, M.J.; Gay, N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982, 1, 945–951. [Google Scholar] [PubMed]
- Schulz, G.E. Binding of nucleotides by proteins. Curr. Opin. Struct. Biol. 1992, 2, 61–67. [Google Scholar] [CrossRef]
- Lanzilotta, W.N.; Ryle, M.J.; Seefeldt, L.C. Nucleotide hydrolysis and protein conformational changes in Azotobacter vinelandii nitrogenase iron protein: Defining the function of aspartate 129. Biochemistry 1995, 34, 10713–10723. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.A.; Mortenson, L.E. Effect of magnesium adenosine 5′-triphosphate on the accessibility of the iron of clostridial azoferredoxin, a component of nitrogenase. Biochemistry 1974, 13, 2382–2388. [Google Scholar] [CrossRef] [PubMed]
- Ljones, T.; Burris, R.H. Nitrogenase: The reaction between iron protein and bathophenanthrolinedisulfonate as a probe for interactions with MgATP. Biochemistry 1978, 17, 1866–1872. [Google Scholar] [CrossRef] [PubMed]
- Deits, T.L.; Howard, J.B. Kinetics of MgATP-dependent iron chelation from the Fe-protein of the Azotobacter vinelandii nitrogenase complex. Evidence for two states. J. Biol. Chem. 1989, 264, 6619–6628. [Google Scholar] [PubMed]
- Anderson, G.L.; Howard, J.B. Reactions with the oxidized iron protein of Azotobacter vinelandii nitrogenase: Formation of a 2Fe center. Biochemistry 1984, 23, 2118–2122. [Google Scholar] [CrossRef] [PubMed]
- Orme-Johnson, W.H.; Hamilton, W.D.; Jones, T.L.; Tso, M.-Y.W.; Burris, R.H.; Shah, V.K.; Brill, W.J. Electron paramagnetic resonance of nitrogenase and nitrogenase components from Clostridium pasteurianum W5 and Azotobacter vinelandii OP. Proc. Natl. Acad. Sci. USA 1972, 69, 3142–3145. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.E.; Lowe, D.J.; Bray, R.C. Studies by electron paramagnetic resonance on the catalytic mechanism of nitrogenase of Klebsiella pneumoniae. Biochem. J. 1973, 135, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, P.A.; Gorelick, N.J.; Münck, E.; Orme-Johnson, W.H. EPR and Mössbauer studies of nucleotide-bound nitrogenase iron protein from Azotobacter vinelandii. J. Biol. Chem. 1987, 262, 14945–14953. [Google Scholar] [PubMed]
- Yates, M.G. Biological Nitrogen Fixation; Stacey, G., Burris, R.H., Evans, H.J., Eds.; Chapman and Hall: New York, NY, USA, 1992; p. 685. [Google Scholar]
- Hausinger, R.P.; Howard, J.B. Thiol reactivity of the nitrogenase Fe-protein from Azotobacter vinelandii. J. Biol. Chem. 1983, 258, 13486–13492. [Google Scholar] [PubMed]
- Chen, L.; Gavini, N.; Tsuruta, H.; Eliezer, D.; Burgess, B.K.; Doniach, S.; Hodgson, K.O. MgATP-induced conformational changes in the iron protein from Azotobacter vinelandii, as studied by small-angle X-ray scattering. J. Biol. Chem. 1994, 269, 3290–3294. [Google Scholar] [PubMed]
- Willing, A.; Howard, J.B. Cross-linking site in Azotobacter vinelandii complex. J. Biol. Chem. 1990, 265, 6596–6599. [Google Scholar] [PubMed]
- Willing, A.H.; Georgiadis, M.M.; Rees, D.C.; Howard, J.B. Cross-linking of nitrogenase components. Structure and activity of the covalent complex. J. Biol. Chem. 1989, 264, 8499–8503. [Google Scholar] [PubMed]
- Wolle, D.; Dean, D.; Howard, J. Nucleotide-iron-sulfur cluster signal transduction in the nitrogenase iron-protein: The role of Asp125. Science 1992, 258, 992–995. [Google Scholar] [CrossRef] [PubMed]
- Seefeldt, L.C. Docking of nitrogenase iron-and molybdenum-iron proteins for electron transfer and MgATP hydrolysis: The role of arginine 140 and lysine 143 of the Azotobacter vinelandii iron protein. Protein Sci. 1994, 3, 2073–2081. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.L.; Davis, L.C.; Rider, M.; Takemoto, D.J. Characterization of nifH mutations of Klebsiella pneumoniae. J. Bacteriol. 1988, 170, 4015–4022. [Google Scholar] [CrossRef] [PubMed]
- Lowery, R.G.; Chang, C.L.; Davis, L.C.; McKenna, M.C.; Stevens, P.J.; Ludden, P.W. Substitution of histidine for arginine-101 of dinitrogenase reductase disrupts electron transfer to dinitrogenase. Biochemistry 1989, 28, 1206–1212. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-Y.; Ledbetter, R.; Shaw, S.; Pence, N.; Tokmina-Lukaszewska, M.; Eilers, B.; Guo, Q.; Pokhrel, N.; Cash, V.L.; Dean, D.R.; et al. Evidence that the Pi release event is the rate-limiting step in the nitrogenase catalytic cycle. Biochemistry 2016, 55, 3625–3635. [Google Scholar] [CrossRef] [PubMed]
- Tezcan, F.A.; Kaiser, J.T.; Howard, J.B.; Rees, D.C. Structural evidence for asymmetrical nucleotide interactions in nitrogenase. J. Am. Chem. Soc. 2015, 137, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Lanzilotta, W.N.; Parker, V.D.; Seefeldt, L.C. Thermodynamics of nucleotide interactions with the Azotobacter vinelandii nitrogenase iron protein. Biochim. Biophys. Acta Protein Struct. Mol. 1999, 1429, 411–421. [Google Scholar] [CrossRef]




| Protein | PDB Code | Resolution (Å) | Reference |
|---|---|---|---|
| NifH | 1G5P | 2.2 | [36] |
| NifH as [Fe4S4]0 “all ferrous” | 1G1M | 2.25 | [36] |
| NifH + MgADP | 1FP6 | 2.15 | [46] |
| ΔL127-NifH + MgATP | 2C8V | 2.5 | [47] |
| NifH + NifDK | 2AFH | 2.1 | [48] |
| ΔL127-NifH + NifDK + MgATP | 1G21 | 3.0 | [3] |
| NifH + NifDK + MgAMPPCP | 4WZB | 2.3 | [48] |
| NifH + NifDK + MgAMP·AlF4− | 1N2C | 3.0 | [49] |
| NifH + NifDK + MgADP | 2AFI | 3.1 | [48] |
| NifH + NifDK + MgADP/MgAMPPCP | 4WZA | 1.9 | [48] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jasniewski, A.J.; Sickerman, N.S.; Hu, Y.; Ribbe, M.W. The Fe Protein: An Unsung Hero of Nitrogenase. Inorganics 2018, 6, 25. https://doi.org/10.3390/inorganics6010025
Jasniewski AJ, Sickerman NS, Hu Y, Ribbe MW. The Fe Protein: An Unsung Hero of Nitrogenase. Inorganics. 2018; 6(1):25. https://doi.org/10.3390/inorganics6010025
Chicago/Turabian StyleJasniewski, Andrew J., Nathaniel S. Sickerman, Yilin Hu, and Markus W. Ribbe. 2018. "The Fe Protein: An Unsung Hero of Nitrogenase" Inorganics 6, no. 1: 25. https://doi.org/10.3390/inorganics6010025
APA StyleJasniewski, A. J., Sickerman, N. S., Hu, Y., & Ribbe, M. W. (2018). The Fe Protein: An Unsung Hero of Nitrogenase. Inorganics, 6(1), 25. https://doi.org/10.3390/inorganics6010025