A Proteomic- and Bioinformatic-Based Identification of Specific Allergens from Edible Insects: Probes for Future Detection as Food Ingredients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Insect Samples
2.1.2. Chemicals
2.2. Edible Insect Protein Extract
2.3. Digestion and Nano-LC-MS/MS Analysis
2.4. Bioinformatics
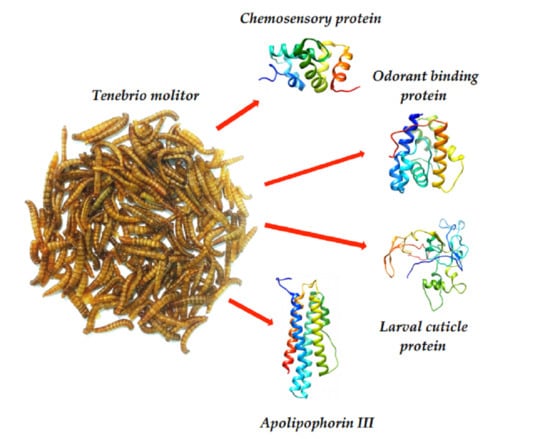
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nadeau, L.; Nadeau, I.; Franklin, F.; Dunkel, F. The potential for entomophagy to address undernutrition. Ecol. Food Nutr. 2015, 54, 200–208. [Google Scholar] [CrossRef] [PubMed]
- van Huis, A. Edible insects are the future? Proc. Nutr. Soc. 2016, 75, 294–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grau, T.; Vilcinskas, A.; Joop, G. Sustainable farming of the mealworm Tenebrio molitor for the production of food and feed. Z. Naturforsch. C J. Biosci. 2017, 72, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Chen, X.M.; Zhao, M.; He, Z.; Sun, L.; Wang, C.Y.; Ding, W.F. Edible insects in China: Utilization and prospects. Insect Sci. 2018, 25, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Raheem, D.; Carrascosa, C.; Bolanle Oluwole, O.; Nieuwland, M.; Saraiva, A.; Millán, R.; Raposo, A. Traditional consumption of and rearing edible insects in Africa, Asia and Europe. Crit. Rev. Food Sci. Nutr. 2019, 59, 2169–2188. [Google Scholar] [CrossRef] [PubMed]
- Fasolin, L.H.; Pereira, R.N.; Pinheiro, A.C.; Martins, J.T.; Andrade, C.C.P.; Ramos, O.L.; Vicente, A.A. Emergent food proteins-towards sustainability, health and innovation. Food Res. Inter. 2019, 125, 108586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlüter, O.; Rumpold, B.A. Insects as food in Europe. J. Insects Food Feed 2019, 5, 1. [Google Scholar] [CrossRef]
- Salter, A.M. Insect proteins: A sustainable and healthy alternative to animal protein? J. Nutr. 2019, 149, 545–546. [Google Scholar] [CrossRef]
- Sorjonem, J.M.; Valtonen, A.; Hirvisalo, E.; Karhapää, M.; Lehtovaara, V.J.; Lindgren, J.; Marnila, P.; Mooney, P.; Mäki, M.; Siljander-Rasi, H.; et al. The plant-based by-product diets for the mass-rearing of Acheta domesticus and Gryllus bimaculatus. PLoS ONE 2019, 14, e0218830. [Google Scholar]
- Pali-Schöll, I.; Binder, R.; Moens, Y.; Polesny, F.; Monsó, S. Edible insects—Defining knowledge gaps in biological and ethical considerations of entomophagy. Crit. Rev. Food Sci. Nutr. 2019, 59, 2760–2771. [Google Scholar] [CrossRef] [Green Version]
- Carcea, M. Quality and nutritional/textural properties of durum wheat pasta enriched with cricket powder. Foods 2020, 9, 1298. [Google Scholar] [CrossRef] [PubMed]
- van Huis, A. Insects as food and feed, a new emerging agricultural sector: A review. J. Insects Food Feed 2020, 6, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, T.; Andere, A.A.; Kelstrup, H.; Emery, V.J.; Picard, C.J. The yellow mealworm (Tenebrio molitor) genome: A resource for the emerging insects as food and feed industry. J. Insects Food Feed 2020, 6, 27–44. [Google Scholar] [CrossRef]
- Jantzen da Silva Lucas, A.; Menegon de Oliveira, L.; da Rocha, M.; Prentice, C. Edible insects: An alternative of nutritional, functional and bioactive compounds. Food Chem. 2020, 311, 126022. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, K.J.; Lopez-Viso, C.; Brameld, J.M.; Parr, T.; Salter, A.M. Insects: A potential source of protein and other nutrients for feed and food. Ann. Rev. Anim. Biosci. 2020, in press. [Google Scholar] [CrossRef]
- Houbraken, M.; Spranghers, T.; De Clercq, P.; Cooreman-Algoed, M.; Couchement, T.; De Clercq, G.; Verbeke, S.; Spanoghe, P. Pesticide comtamination of Tenebrio molitor (Coleoptera: Tenebrionidae) for human consumption. Food Chem. 2016, 201, 264–269. [Google Scholar] [CrossRef]
- Schlüter, O.; Rumpold, B.; Holzhauser, T.; Roth, A.; Vogel, R.F.; Quasigroch, W.; Vogel, S.; Heinz, V.; Jäger, H.; Bandick, N.; et al. Safety aspects of the production of foods and food ingredients from insects. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef]
- Testa, M.; Stillo, M.; Maffei, G.; Androlo, V.; Gardois, P.; Zotti, C.M. Ugly but tasty: A systematic review of possible human and animal health risks related to entomophagy. Crit. Rev. Food. Sci. Nutr. 2017, 57, 3747–3759. [Google Scholar] [CrossRef]
- Poma, G.; Cuykx, M.; Amato, C.; Calaprice, C.; Focant, J.-F.; Covaci, A. Evaluation of hazardous chemicals in edible insects and insect-based food intended for human consumption. Food Chem. Toxicol. 2017, 100, 70–79. [Google Scholar] [CrossRef]
- Montowska, M.; Kowalczewski, P.L.; Rybicka, I.; Fornal, E. Nutritional value, protein and peptide composition of edible cricket powders. Food Chem. 2019, 289, 130–138. [Google Scholar] [CrossRef]
- Poma, G.; Yin, S.; Tang, B.; Fujii, Y.; Cuykx, M.; Covaci, A. Occurrence of selected organic comtaminants in edible insects and assessment of their chemical safety. Environ. Health Perspect. 2019, 127, 127009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truzzi, C.; Illuminati, S.; Girolametti, F.; Antonucci, M.; Scarponi, G.; Ruschioni, S.; Riolo, P.; Annibaldi, A. Influence of feeding substrates on the presence of toxic metals (Cd, Pb, Ni, As, Hg) in larvae of Tenebrio molitor: Risk assessment for human consumption. J. Environ. Res. Public Health 2019, 16, 4815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garofalo, C.; Osimani, A.; Milanović, V.; Taccari, M.; Cardinali, F.; Aquilanti, L.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Clementi, F. The microbiota of marketed processed edible insects as revealed by high-throughput sequencing. Food Microbiol. 2017, 62, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Wynants, E.; Crauwels, S.; Verreth, C.; Gioanotten, N.; Lievens, B.; Claes, J.; Van Campenhout, L. Microbial dynamics during production of lesser mealworms (Alphitobius diaperinus) for human consumption at industrial scale. Food Microbiol. 2018, 70, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Wynants, E.; Frooninckx, L.; Crauwels, S.; Verreth, C.; De Smet, J.; Sandrock, C.; Wohlfahrt, J.; Van Schelt, J.; Depraetere, S.; Lievens, B.; et al. Assessing the microbiota of black soldier fly larvae (Hermetia illucens) reared on organic waste streams on four different locations at laboratory and large scale. Microbiol. Ecol. 2019, 77, 913–930. [Google Scholar] [CrossRef]
- Gałęcki, R.; Sokół, R. A parasitological evaluation of edible insects and their role in the transmission of parasitic diseases to humans and animals. PLoS ONE 2019, 14, e0219303. [Google Scholar] [CrossRef] [Green Version]
- Barre, A.; Caze-Subra, S.; Gironde, C.; Bienvenu, F.; Bienvenu, J.; Rougé, P. Entomophagy and the risk of allergy. Rev. Fr. Allergol. 2014, 54, 315–321. [Google Scholar] [CrossRef]
- Belluco, S.; Losasso, C.; Maggioletti, M.; Alonzi, C.; Ricci, A.; Paoletti, M.G. Edible insects: A food security solution or a food safety concern? Anim. Front. 2015, 5, 25–30. [Google Scholar]
- Verhoeckx, K.; Broekman, H.; Knulst, A.; Houben, G. Allergenicity assessment strategy for novel food proteins and protein sources. Regul. Toxicol. Pharmacol. 2016, 79, 118–124. [Google Scholar] [CrossRef]
- Quirce, S.; Antolín-Amérigo, D.; Domínguez-Ortega, J. Hidden occupational allergens such as additives. Curr. Opin. Allergy Clin. Immunol. 2018, 18, 67–72. [Google Scholar] [CrossRef]
- de Gier, S.; Verhoeckx, K. Insect (food) allergy and allergens. Mol. Immunol. 2018, 100, 82–106. [Google Scholar] [CrossRef] [PubMed]
- Lee, Q.C.; Kim, S.R.; Park, K.H.; Lee, J.H.; Park, J.W. Clinical features and culprit food allergens of Korean adult food allergy patients: A cross-sectional single-institute study. Allergy Asthma Immunol. Res. 2019, 11, 723–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, K.Y.; Park, J.W. Insect allergens on the dining table. Curr. Protein Pept. Sci. 2020, 21, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, P.; Courtois, J.; Van der Brempt, X.; Tollenaere, S. Food-induced anaphylaxis to Tenebrio molitor and allergens implicated. Rev. Fr. Allergol. 2019, 59, 389–393. [Google Scholar] [CrossRef]
- Gautreau, M.; Restuccia, M.; Senser, K.; Weisberg, S.N. Familial anaphylaxis after silkworm ingestion. Prehosp. Emerg. Care 2017, 21, 83–85. [Google Scholar] [CrossRef]
- Verhoeckx, K.; van Broekhoven, S.; den Hartog-Jager, C.F.; Gaspari, M.; de Jong, G.A.; Wichers, H.J.; van Hoffen, E.; Houben, G.F.; Knulst, A.C. House dust mite (Der p 10) and crustacean allergic patients may react to food containing yellow mealworm proteins. Food Chem. Toxicol. 2014, 65, 364–373. [Google Scholar] [CrossRef]
- Broekman, H.; Verhoeckx, K.; den Hartog Jager, C.F.; Kuizinga, A.G.; Pronk-Kleinjan, M.; Remington, B.C.; Bruijnzeel-Koomen, C.A.; Houben, G.F.; Knulst, A.C. Majority of shrimp-allergic patients are allergic to mealworm. J. Allergy Clin. Immunol. 2016, 137, 1261–1263. [Google Scholar] [CrossRef] [Green Version]
- Broekman, H.C.H.; Knulst, A.; de Jong, G.; Gaspari, M.; den Hartog Jager, C.F.; Houben, G.H.; Verhoeckx, K.C. Is mealworm or shrimp allergy indicative for food allergy to insects? Mol. Nutr. Food Res. 2017, 61, 1601061. [Google Scholar] [CrossRef]
- Barre, A.; Simplicien, M.; Cassan, G.; Benoist, H.; Rougé, P. Food allergen families common to different arthropods (mites, insects, crustaceans), mollusks and nematods: Cross-reactivity and potential cross-allergenicity. Rev. Fr. Allergol. 2018, 58, 581–593. [Google Scholar] [CrossRef]
- Kamemura, N.; Sugimoto, M.; Tamehiro, N.; Adachi, R.; Tomonari, S.; Watanabe, T.; Mito, T. Cross-allergenicity of crustacean and the edible insect Gryllus bimaculatus in patients with shrimp allergy. Mol. Immunol. 2019, 106, 127–134. [Google Scholar] [CrossRef]
- Francis, F.; Doyen, V.; Debaugnies, F.; Mazzucchelli, G.; Caparros, R.; Alabi, T.; Blecker, C.; Haubruge, E.; Corazza, F. Limited cross reactivity among arginine kinase allergen from mealworm and cricket edible insects. Food Chem. 2019, 276, 714–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pali-Schöll, I.; Meinlschmidt, P.; Larenas-Linnemann, D.; Purschke, B.; Hofstetter, G.; Rodríguez-Monroy, F.A.; Einhorn, L.; Mothes-Luksch, N.; Jensen-Jarolim, E.; Jäger, H. Edible insects: Cross-recognitionof IgE from crustacean- and house dust mite allergic patients, and reduction of allergenicity by food processing. World Allergy Organ. J. 2019, 12, 100006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, L.; Hoang, J.A.; Kothari, A.; Eiwegger, T.; Vadas, P. Shellfish allergy is a risk factor for cricket anaphylaxis. J. Allergy Clin. Immunol. Pract. 2020, 8, 2396–2398. [Google Scholar] [CrossRef] [PubMed]
- Palmer, L.K.; Marsh, J.T.; Lu, M.; Goodman, R.E.; Zeece, M.G.; Johnson, P.E. Shellfish tropomyosin IgE cross-reactivity differs among edible insect species. Mol. Nutr. Food Res. 2020, 64, e1900923. [Google Scholar] [CrossRef] [PubMed]
- Tramuta, C.; Gallina, S.; Bellio, A.; Bianchi, D.M.; Chiesa, F.; Rubiola, S.; Romano, A.; Decastelli, L. A set of multiplex polymerase chain reactions for genomic detection of nine edible insect species in foods. J. Insect Sci. 2018, 18, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Barre, A.; Pichereaux, C.; Valazquez, E.; Maudouit, A.; Simplicien, M.; Garnier, L.; Bienvenu, F.; Bienvenu, J.; Burlet-Schiltz, O.; Auriol, C.; et al. Insights into the allergenic potential of the edible yellow mealworm (Tenebrio molitor). Foods 2019, 8, 515. [Google Scholar] [CrossRef] [Green Version]
- Bouyssié, D.; Hesse, A.M.; Mouton-Barbosa, E.; Rompais, M.; Macron, C.; Carapito, C.; Gonzalez de Peredo, A.; Dupierris, V.; Burel, A.; Menetrey, J.P.; et al. Proline: An efficient and user-friendly software suite for large-scale proteomics. Bioinformatics 2020, 36, 3148–3155. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 1999, 15, 4876–4882. [Google Scholar]
- Risler, J.-L.; Delorme, M.O.; Delacroix, H.; Henaut, A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new an efficient scoring matrix. J. Mol. Biol. 1998, 204, 1019–1029. [Google Scholar] [CrossRef]
- Breiter, D.R.; Kanost, M.R.; Benning, M.M.; Wesenberg, G.; Law, J.H.; Wells, M.A.; Rayment, I.; Holden, H.M. Molecular structure of an apolipoprotein determined at 2.5-Å resolution. Biochemistry 1991, 30, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Zheng, Y.; Yang, D.; Wang, J. NMR solution structure and dynamics of an exchangeable apolipoprotein, Locusta migratoria apolipophorin III. J. Biol. Chem. 2003, 278, 21212–21220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, S.; Chmelik, J.; Zidek, L.; Padrta, P.; Novák, P.; Zdráhal, Z.; Picimbon, J.F.; Löfstedt, C.; Sklenár, V. Structure of Bombyx mori chemosensory protein 1 in solution. Arch. Insect Biochem. Physiol. 2007, 66, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, S.; Liou, Y.C.; Davies, P.L.; Krause, E.; Sönnichsen, F.D. A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands. Structure 1999, 7, 1325–1332. [Google Scholar] [CrossRef] [Green Version]
- Spinelli, S.; Lagarde, A.; Iovinella, P.; Legrand, P.; Tegoni, M.; Pelosi, P.; Cambillau, C. Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem. Mol. Biol. 2012, 42, 41–50. [Google Scholar] [CrossRef]
- Zhou, J.J.; Robertson, G.; He, X.; Dufour, S.; Hooper, A.M.; Pickett, J.A.; Keep, N.H.; Field, L.M. Characterisation of Bombyx mori odorant-binding proteins reveals that a general odorant-binding protein discriminates between sex pheromone components. J. Mol. Biol. 2009, 389, 529–545. [Google Scholar] [CrossRef] [Green Version]
- Sandler, B.H.; Nikonova, L.; Leal, W.S.; Clardy, J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein-bombykol complex. Chem. Biol. 2000, 7, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Pietrzyk, A.; Bujacz, A.; Mueller-Dieckmann, J.; Łochynska, M.; Jalkolski, M.; Bujacz, G. Crystallographic identification of an unexpected protein complex in silkworm haemolymph. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 2353–2364. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA-a self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Ryu, K.S.; Lee, J.O.; Kwon, T.H.; Choi, H.H.; Park, H.S.; Hwang, S.K.; Lee, Z.W.; Lee, K.B.; Han, Y.H.; Choi, Y.S.; et al. The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin. Biochem. J. 2009, 421, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Baba, S.; Matsuo, K.; Ito, S.; Mikami, B. The high-resolution crystal structure of lobster hemocyanin shows its enzymatic capability as a phenoloxidase. Arch. Biochem. Biophys. 2020, 688, 108370. [Google Scholar] [CrossRef] [PubMed]
- Lagarde, A.; Spinelli, S.; Tegoni, M.; He, X.; Field, L.; Zhou, J.J.; Cambillau, C. The crystal structure of odorant binding protein 7 from Anopheles gambiae exhibits an outstanding adaptability of its binding site. J. Mol. Biol. 2011, 414, 401–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- di Luccio, E.; Ishida, Y.; Leal, W.S.; Wilson, D.K. Crystallographic observation of pH-induced conformational changes in the Amyelois transitella pheromone-binding protein AtraPBP1. PLoS ONE 2013, 8, e53840. [Google Scholar] [CrossRef] [Green Version]
- Damberger, F.F.; Ishida, Y.; Leal, W.S.; Wüthrich, K. Structural basis of ligand binding and release in insect pheromone-binding protein: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. J. Mol. Biol. 2007, 373, 811–819. [Google Scholar] [CrossRef]
- Jímenez-Sandoval, P.; Madrigal-Carrillo, E.A.; Santamaría-Suárez, H.A.; Maturana, D.; Rentería-González, I.; Benitez-Cardoza, C.G.; Torres-Larios, A.; Brieba, L.G. Mimicking a p53-MDM2 interaction based on a stable immunoglobulin-like domain scaffold. Proteins 2018, 86, 802–812. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemistry of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Melo, F.; Feytmans, E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998, 277, 1141–1152. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M. Meng, E.C.; Ferrin, T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Human Press Inc.: Totowa, NJ, USA, 2005; Volume 52, pp. 571–607. [Google Scholar]
- Pelosi, P.; Calvello, M.; Ban, L. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem. Senses 2005, 30, i291–i292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khurana, T.; Collison, M.; Chew, F.T.; Slater, J.E. Bla g 3: A novel allergen of German cockroach identified using cockroach-specific avian single-chain variable fragment antibody. Ann. Allergy Asthma Immunol. 2014, 112, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Lee, M.F.; Liao, S.C.; Luo, S.F. Sequencing analysis of cDNA clones encoding the American cockroach Cr-PI allergens. Homology with insect hemolymph proteins. J. Biol. Chem. 1996, 271, 17937–17943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinroch, C.; Srisomsap, C.; Chokchaichamnankit, D.; Punyarit, P.; Phiriyangkul, P. Identification of novel allergen in edible insect, Gryllus bimaculatus and its cross-reactivity with Macrobrachium spp. allergens. Food Chem. 2015, 184, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Just, N.; Lièvre, K.; Lallemand, K.; Leduc, V. Implication de l’hexamérine dans un cas d’allergie aux larves de pouches. Rev. Fr. Allergol. 2012, 52, 258. [Google Scholar] [CrossRef]
- Colomb, S.; Bourrain, J.L.; Leduc, V.; Burmester, T.; Marin, G.; Lesage, F.X.; Dhivert-Donnadieu, H. Demoly, P. Identification of the larval serum proteins as major fruit fly (Drosophila melanogaster) occupational allergens. J. Allergy Clin. Immunol. Pract. 2017, 5, 1153–1155. [Google Scholar] [CrossRef]
- Broekman, H.C.H.P.; Knulst, A.C.; de Jong, G.; Gaspari, M.; den Hartog Jager, C.F.; Houben, G.F.; Verhoeckx, K.C.M. Primary respiratory and food allergy to mealworm. J. Allergy Clin. Immunol. 2017, 140, 600–603. [Google Scholar] [CrossRef] [Green Version]
- Tomiya, N.; Narang, S.; Lee, Y.C.; Betenbaugh, M.J. Comparing N-glycan processing in mammalian cell lines to native and engineered lepidopteran insect cell lines. Glycoconj. J. 2004, 21, 343–360. [Google Scholar] [CrossRef]
- Scheys, F.; Van Damme, E.J.M.; De Schutter, K.; Staes, A.; Gevaert, K.; Smagghe, G. Evolutionarily conserved and species-specific glycoproteins in the N-glycoproteomes of diverse insect species. Insect Biochem. Mol. Biol. 2018, 100, 22–29. [Google Scholar] [CrossRef]
- van Broekhoven, S.; Bastiaan-Net, S.; de Jong, N.W.; Wichers, H.J. Influence of processing and in vitro digestion on the allergic cross-reactivity of three mealworm species. Food Chem. 2015, 196, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Broekman, H.; Knulst, A.; den Hartog Jager, S.; Monteleone, F.; Gaspari, M.; de Jong, G.; Houben, G.; Verhoeckx, K. Effect of thermal processing on mealworm allergenicity. Mol. Nutr. Food Res. 2015, 59, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Hall, F.; Johnson, P.E.; Liceaga, A. Effect of enzymatic hydrolysis on bioactive properties and allergenicity of cricket (Gryllodes sigillatus) protein. Food Chem. 2018, 262, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Frigerio, J.; Agostinetto, G.; Sandionigi, A.; Mezzasalma, V.; Berterame, N.M.; Casiraghi, M.; Labra, M.; Galimberti, A. The hidden ‘plant side’ of insect novel foods: A DNA-based assessment. Food. Res. Int. 2020, 128, 108751. [Google Scholar] [CrossRef] [PubMed]
- Klueber, J.; Costa, J.; Randow, S.; Codreanu-Morel, F.; Verhoeckx, K.; Bindslev-Jensen, C.; Ollert, M.; Hoffmann-Sommergruber, K.; Morisset, M.; Holzhauser, T.; et al. Homologous tropomyosins from vertebrate and invertebrate: Recombinant calibrator proteins in functional biological assays for tropomyosin allergenicity assessment of novel animal foods. Clin. Exp. Allergy 2020, 50, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Nebbia, S.; Lamberti, C.; Giorgis, V.; Giuffrida, M.G.; Manfredi, M.; Marengo, E.; Pessione, E.; Schiavone, A.; Boita, M.; Brussino, L.; et al. The cockroach allergen-like protein is involved in primary respiratory and food allergy to yellow mealworm (Tenebrio molitor). Clin. Exp. Allergy 2019, 49, 1379–1382. [Google Scholar] [CrossRef]
- Raheem, D.; Raposo, A.; Oluwole, O.B.; Nieuwland, M.; Saraiva, A.; Carrascosa, C. Entomophagy: Nutritional, ecological, safety and legislation aspects. Food Res. Intern. 2019, 126, 108672. [Google Scholar] [CrossRef]
- Govorushko, S. Global status of insects as food and feed source: A review. Trends Food Sci. Technol. 2019, 91, 436–445. [Google Scholar] [CrossRef]
- Oonincx, D.G.A.B.; de Boer, I.J.M. Environmental impact of the production of mealworms as a protein source for humans—A life cycle assessment. PLoS ONE 2012, 7, e51145. [Google Scholar] [CrossRef] [Green Version]
- Lundy, M.E.; Parrella, M.P. Crickets are not a free lunch: Protein capture from scalable organic side-streams via high-density populations of Acheta domesticus. PLoS ONE 2015, 10, e0118785. [Google Scholar] [CrossRef]
- Rumpold, B.A.; Schlüter, O.K. Potential and challenges of insects as an innovative source for food and feed production. Innov. Food Sci. Emerging Technol. 2013, 17, 1–11. [Google Scholar] [CrossRef]
- Payne, C.L.R.; Dobermann, D.; Forkes, A.; House, J.; Josephs, J.; McBride, A.; Müller, A.; Quilliam, R.S.; Soares, S. Insects as food and feed: European perspectives on recent research and future priorities. J. Insects Food Feed 2016, 2, 269–276. [Google Scholar] [CrossRef] [Green Version]
- Gmuer, A.; Nuessli Gith, J.; Hartmann, C.; Siegrist, M. Effects of the degree of processing of insect ingredients in snacks on expected emotional experiences and willingness to eat. Food Quality Prefer. 2016, 54, 117–127. [Google Scholar] [CrossRef]
- House, J. Consumer acceptance of insect-based foods in the Netherlands: Academic anf commercial implications. Apetite 2016, 107, 47–58. [Google Scholar] [CrossRef] [Green Version]
- La Barbera, F.; Verneau, F.; Amato, M.; Grunert, K. Understanding Westerners’ disgust for the eating of insects: The role of food neophobia and implicit associations. Food Quality Prefer. 2018, 64, 120–125. [Google Scholar] [CrossRef]
- Woolf, E.; Zhu, Y.; Emory, K.; Zhao, J.; Liu, C. Willingness to consume insect-containing foods: A survey in the United States. LWT 2019, 102, 100–105. [Google Scholar] [CrossRef]
- EFSA Scientific Committee. Scientific Opinion on a risk profile related to production and consumption of insects as food and feed. EFSA J. 2015, 13, 4257. [Google Scholar] [CrossRef] [Green Version]
- EFSA NDA Panel (EFSA Panel on Nutrition, Novel Foods and Food Allergens); Turck, D.; Castenmiller, J.; De Henauw, S.; Hirsch-Ernst, K.I.; Kearney, J.; Maciuk, A.; Mangelsdorf, I.; McArdle, H.J.; Naska, A.; et al. Scientific Opinion on the safety of dried yellow mealworm (Tenebrio molitor larva) as a novel food pursuant to Regulation (EU) 2015/2283. EFSA J. 2021, 19, 6343. [Google Scholar]










| 1. Sex specific storage protein 1 | 55. Mitochondrial aldehyde dehydrogenase | cysteine |
| 2. Arylphorin I | 56. Hemocytin | 108. Paralytic peptide binding protein |
| 3. Sex specific storage protein 2 | 57. Antichymotrypsin-1 | 109. ATP-dependent (S)-NAD(P)H-hydrate |
| 4. Silkworm storage protein | 58. Pyruvate kinase | dehydratase |
| 5. Arylphorine 2 | 59. Fructose-1,6-biphosphatase | 110. Chymotrypsin inhibitor fb |
| 6. Apolipophorin | 60. 30 kDa protein | 111. 30K protein 14 |
| 7. Vitellogenin | 61. Chemosensory protein 7 | 112. Glucosamine-6-phosphate isomerase |
| 8. Vitellogenin | 62. Nucleoside diphosphate kinase | 113. Thioredoxin |
| 9. Antitrypsin isoform 1 | 63. Thiol peroxiredoxin | 114. Adenosylhomocysteinase |
| 10. Aliphatic nitrilase | 64. Tropomyosin I | 115. Proteasome subunit α type |
| 11. Antichymotrypsin-2 | 65. Promoting protein | 116. Carboxypeptidase |
| 12. 30K protein 3 | 66. Phosphatidylethanolamine binding | 117. Immune-related protein 1 |
| 13. Hemolin | protein isoform 2 | 118. Glutathione S-transferase δ |
| 14. 30K protein 7 | 67. Obstructor-A | 119. Heat shock protein 70-3 |
| 15. Low molecular 30 kDa lipoprotein | 68. Mitochondrial aldehyde dehydrogenase | 120. 30K protein 13 |
| 16. Hemolin | 69. Actin-1 | 121. Heat shock cognate 70 protein |
| 17. Major plasma protein 30K | 70. Mesencephalic astrocyte-derived | 122. DJ-1 β |
| 18. Hemolin | neurotrophic factor | 123. Chemosensory protein 9 |
| 19. Chitooligosaccharidolytic β-N-acetyl- | 71. Malate dehydrogenase | 124. Allergen |
| glucosaminidase | 72. Transaldolase | 125. 14-3-3 protein ζ |
| 20. Odorant binding protein | 73. Malic enzyme | 126. 32 kDa apolipoprotein |
| 21. Odorant-binding protein 6 | 74. Glyceraldehyde-3-phosphate | 127. Carboxypeptidase inhibitor |
| 22. Aliphatic nitrilase | dehydrogenase | 128. Lysozyme |
| 23. Dihydrolipoyl dehydrogenase | 75. Chitinase 3 | 129. tRNA-nucleotidyltransferase 1 |
| 24. 30K protein 4 | 76. Chemosensory protein-1 | 130. Serpin-11 |
| 25. Carboxylic ester hydrolase | 77. Serine hydroxymethyltransferase | 131. Myosin heavy chain, non-muscle |
| 26. 27 kDa glycoprotein | 78. Acyl-CoA binding protein | 132. Heat shock 70 kDa protein cognate 3 |
| 27. Carboxylic ester hydrolase | 79. Isocitrate dehydrogenase [NADP] | 133. Chemosensory protein 8 |
| 28. Tropomyosin-1 isoform 2 | 80. Cuticle protein | 134. Fructose-1,6-biphosphatase 1 |
| 29. Low molecular mass 34 kDa lipoprotein | 81. Cationic peptide CP8 | 135. Ubiquitin |
| 21G1 | 82. Tropomyosin-1 isoforms 33/34 | 136. N-acetylglucosamine-6-phosphate |
| 30. Putative peptidase | 83. β-galactosidase | deacetylase |
| 31. Saposin-like protein | 84. Odorant binding protein | 137. Mitochondrial cytochrome C |
| 32. Hydroxypyruvate isomerase | 85. Glyceraldehyde-3-phosphate | 138. Pro-phenol oxidase |
| 33. Serpin-2 | dehydrogenase | 139. Catalase |
| 34. Serpin-6 | 86. Tenebrin | 140. Glutathione-S-Transferase 1 |
| 35. Glucosamine-6-phosphate isomerase | 87. Aminoacylase | 141. Thioredoxin peroxidase |
| 36. Serpin-9 | 88. ARP-like protein | 142. Peroxiredoxin 1 |
| 37. Serpin-2 | 89. Serpin-7 | 143. Glutamate dehydrogenase |
| 38. Hemolymph juvenile hormone binding | 90. Serpin-3 | 144. Nucleoplasmin-like protein |
| protein | 91. Calmodulin | 145. Angiotensin converting enzyme |
| 39. Molting fluid carboxypeptidase A | 92. Imaginal disk growth factor | 146. DNA supercoiling factor |
| 40. Superoxide dismutase [Cu-Zn] | 93. Actin-depolymerizing factor 1 | 147. Nucleoside diphosphate kinase |
| 41. Fructose-biphosphate aldolase | 94. Chymotrypsin inhibitor SCI-III | 148. HSP70 |
| 42. Fibrillin-like protein | 95. Myosin light chain 2 | 149. Sericin 2 |
| 43. Bm8 interacting protein 2d-4 | 96. POX-C | 150. Molting carboxypeptidase A |
| 44. Superoxide dismutase [Cu-Zn] | 97. Scarface | 151. Fibrillin-1 |
| 45. Type IV collagen | 98. Proteasome subunit α type | 152. Small heat shock protein 20.8 |
| 46. Glyceraldehyde-3-phosphate | 99. Chymotrypsin inhibitor SCI-I | 153. β-glucuronidase |
| dehydrogenase | 100. Kazal-type proteinase inhibitor | 154. Trehalase |
| 47. Serpin-5 | 101. Polyubiquitin-c isoform x7 | 155. DNA (apurinic or apyrimidinic site) |
| 48. Superoxide dismutase [Cu-Zn] | 102. Chymotrypsin inhibitor SCI-II | lyase |
| 49. Actin | 103. Carboxylic ester hydrolase | 156. Cuticlin-1 |
| 50. Thioredoxin | 104. Prophenoloxidase subunit 2 | 157. Kv-channel-interacting protein |
| 51. Glucosamine-6-phosphate isomerase | 105. Thiol peroxiredoxin | 158. Ferritin |
| 52. Cystathionine γ-lyase | 106. Ubiquitin/ribosomal protein S27 Ae | 159. Chemosensory protein 5 |
| 53. Retinoic acid binding protein | fusion protein | 160. Arylphorin subunit α |
| 54. Putative actin-related protein | 107. Secreted protein acidic and rich in | 161. Pterin carbinolamin dehydratase |
| Allergen | Bm | Tm | Ad | Lm | Zm | Rf | C | Ac | I | M | W | F | P | An |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acidic ribosomal protein | + | + | + | X | X | X | X | X | X | X | X | |||
| Actin α | + | + | + | + | + | + | X | X | X | X | X | X | X | X |
| Actinin | + | + | X | X | X | X | X | X | X | X | ||||
| Adenosylhomocysteinase | + | X | X | X | ||||||||||
| α-Amylase | + | X | X | X | X | X | X | X | X | |||||
| Apolipophorin III | + | + | + | X | X | X | X | X | X | |||||
| Apolipoprotein | + | X | X | X | X | X | X | X | X | |||||
| Arginine kinase | + | + | + | + | + | + | X | X | X | X | X | X | ||
| Arylphorin, Hemocyanin | + | + | + | + | X | X | ||||||||
| Aspartic protease | + | X | X | X | X | X | X | |||||||
| ATP synthase | + | + | + | X | X | X | X | X | X | X | X | |||
| Carboxypeptidase | + | X | X | X | X | X | X | X | X | |||||
| Catalase | + | X | X | X | X | X | X | X | X | |||||
| Chemosensory protein | + | + | + | X | X | X | X | X | ||||||
| Chitinase | + | + | X | X | X | X | X | X | X | X | ||||
| Cockroach allergen-like protein | + | X | ||||||||||||
| Cystatin proteinase inhibitor | + | X | X | X | X | X | X | X | X | |||||
| Cytochrome C | + | + | + | X | X | X | X | X | X | X | X | |||
| Enolase | + | + | + | + | + | X | X | X | X | X | X | X | X | |
| Fatty acid-binding protein | + | + | + | + | X | X | X | X | X | X | X | X | ||
| Ferritin | + | X | X | X | X | X | X | X | X | |||||
| Fructose-1,6-biphosphate aldolase | + | + | + | + | + | + | X | X | X | |||||
| Glucosamine-6-phosphate isomerase | + | X | X | X | X | X | X | X | X | |||||
| Glutathione S-transferase | + | + | X | X | X | X | X | X | X | X | ||||
| Glyceraldehyde-3-phosphate dehydrogenase | + | + | + | + | + | + | X | X | X | X | X | X | X | X |
| Hexamerin | + | + | + | + | + | X | X | X | ||||||
| HSP 70 | + | + | + | + | + | X | X | X | X | X | X | X | X | |
| Larval cuticle protein | + | + | + | X | X | X | X | X | ||||||
| Lipocalin | + | X | X | X | X | X | X | X | X | |||||
| Lysosomal aspartic protein | + | X | X | X | X | X | X | X | X | |||||
| Lysozyme | + | X | X | X | X | X | X | X | X | |||||
| Malate dehydrogenase | + | + | + | X | X | X | X | X | X | X | X | |||
| Mitochondrial aldehyde dehydrogenase | + | X | X | X | X | X | X | X | X | |||||
| α-Myosin | + | + | + | + | + | X | X | |||||||
| Myosin heavy chain | + | + | X | X | X | X | X | X | X | X | ||||
| Myosin light chain | + | + | X | X | X | X | X | X | X | X | ||||
| Nucleoside diphosphate kinase | + | X | X | X | X | X | X | X | X | |||||
| Odorant-binding protein | + | + | + | + | + | + | X | X | X | X | X | X | ||
| Paramyosin long form | + | + | X | X | X | X | X | X | ||||||
| Paramyosin short form | + | + | X | X | X | X | X | X | X | |||||
| Peroxiredoxin | + | + | X | X | X | X | X | X | X | X | ||||
| Pyruvate kinase | + | + | + | X | X | X | X | X | X | X | X | |||
| Receptor for activated protein kinase | + | X | X | X | X | X | X | X | X | |||||
| Sarcoplasmic calcium-binding protein | + | X | X | X | X | X | X | X | ||||||
| Serine protease | + | X | X | X | X | X | X | X | X | |||||
| Serpin | + | + | X | X | X | X | X | X | X | X | ||||
| Superoxide dismutase [Cu-Zn] | + | + | + | X | X | X | X | X | X | X | X | |||
| Thioredoxin | + | + | X | X | X | X | X | X | X | X | ||||
| Transaldolase | + | X | X | X | X | X | X | X | X | |||||
| Triosephosphate isomerase | + | + | X | X | X | X | X | X | X | X | ||||
| Tropomyosin 1 | + | + | + | + | + | + | X | X | X | X | X | X | X | X |
| Tropomyosin 2 | + | + | + | X | X | X | X | X | X | X | X | |||
| Troponin C | + | + | X | X | X | X | X | X | X | X | ||||
| Troponin T | + | + | + | X | X | X | X | X | X | X | X | |||
| Trypsin | + | X | X | X | X | X | X | X | X | |||||
| Tubulin α | + | + | X | X | X | X | X | X | X | X | ||||
| Tubulin β | + | + | + | + | X | X | X | X | X | X | X | X | ||
| Vitellogenin | + | + | + | X | X | X | X | X | X | X | X |
| Protein: | Insects: | Crustaceans: | Mollusks: | Nematods: |
|---|---|---|---|---|
| Apolipophorin III | 549 (79.7%) | 6 (0.8%) | 26 (3.8%) | 108 (15.7%) |
| Chemosensory protein | 17,207 (98%) | 142 (0.8%) | 47 (0.2%) | 180 (1%) |
| Cockroach allergen-like protein | 1 | N/I | N/I | N/I |
| Hexamerin | 395 (99.5%) | 2 (0.5%) | N/I | N/I |
| Larval culticle protein | 3971 (84.7%) | 315 (6.7%) | 1 (0.02%) | 399 (8.5%) |
| Odorant binding protein | 14,318 (99.8%) | 13 (0.08%) | N/I | 19 (0.13%) |
| Receptor for activated protein kinase | 313 (79%) | 16 (4%) | 17 (4.4%) | 50 (12.6%) |
| Protein: | Bombyx mori | Locusta migratoria | Tenebrio molitor |
|---|---|---|---|
| Chemosensory protein | 16 | 28 | 12 |
| Odorant binding protein | 23 | 26 | 19 |
| Hexamerin | 0 | 7 | 2 |
| Apolipophorin III | 3 | 6 | 1 |
| Larval culticle protein | 9 | 3 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barre, A.; Pichereaux, C.; Simplicien, M.; Burlet-Schiltz, O.; Benoist, H.; Rougé, P. A Proteomic- and Bioinformatic-Based Identification of Specific Allergens from Edible Insects: Probes for Future Detection as Food Ingredients. Foods 2021, 10, 280. https://doi.org/10.3390/foods10020280
Barre A, Pichereaux C, Simplicien M, Burlet-Schiltz O, Benoist H, Rougé P. A Proteomic- and Bioinformatic-Based Identification of Specific Allergens from Edible Insects: Probes for Future Detection as Food Ingredients. Foods. 2021; 10(2):280. https://doi.org/10.3390/foods10020280
Chicago/Turabian StyleBarre, Annick, Carole Pichereaux, Mathias Simplicien, Odile Burlet-Schiltz, Hervé Benoist, and Pierre Rougé. 2021. "A Proteomic- and Bioinformatic-Based Identification of Specific Allergens from Edible Insects: Probes for Future Detection as Food Ingredients" Foods 10, no. 2: 280. https://doi.org/10.3390/foods10020280
APA StyleBarre, A., Pichereaux, C., Simplicien, M., Burlet-Schiltz, O., Benoist, H., & Rougé, P. (2021). A Proteomic- and Bioinformatic-Based Identification of Specific Allergens from Edible Insects: Probes for Future Detection as Food Ingredients. Foods, 10(2), 280. https://doi.org/10.3390/foods10020280