An Untargeted Metabolomics Investigation of Milk from Dairy Cows with Clinical Mastitis by 1H-NMR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Metabolome Analysis
2.3. Statistical Analysis
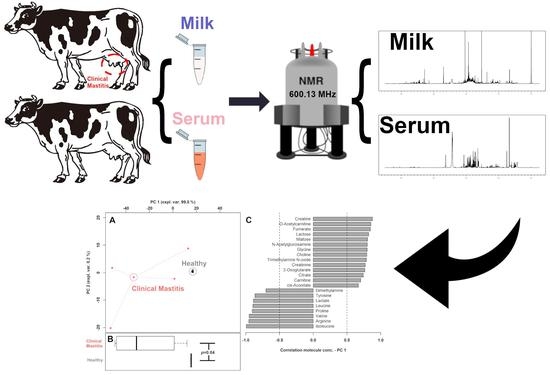
3. Results
3.1. 1H-NMR Spectra of Milk Samples
3.2. Milk Metabolome Affected by Clinical Mastitis
3.3. Pathway Analysis of Serum Metabolites Affected by Clinical Mastitis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Conner, J.G.; Eckersall, P.D.; Doherty, M.; Douglas, T.A. Acute phase response and mastitis in the cow. Res. Vet. Sci. 1986, 41, 126–128. [Google Scholar] [CrossRef]
- Nyman, A.K.; Persson Waller, K.; Bennedsgaard, T.W.; Larsen, T.; Emanuelson, U. Associations of udder-health indicators with cow factors and with intramammary infection in dairy cows. J. Dairy Sci. 2014, 97, 5459–5473. [Google Scholar] [CrossRef] [Green Version]
- Luangwilai, M.; Duangmal, K.; Chantaprasarn, N.; Settachaimongkon, S. Comparative metabolite profiling of raw milk from subclinical and clinical mastitis cows using 1H-NMR combined with chemometric analysis. Int. J. Food Sci. Technol. 2021, 56, 493–503. [Google Scholar] [CrossRef]
- Sargeant, J.M.; Leslie, K.E.; Shirley, J.E.; Pulkrabek, B.J.; Lim, G.H. Sensitivity and Specificity of Somatic Cell Count and California Mastitis Test for Identifying Intramammary Infection in Early Lactation. J. Dairy Sci. 2001, 84, 2018–2024. [Google Scholar] [CrossRef]
- Laghi, L.; Picone, G.; Capozzi, F. Nuclear magnetic resonance for foodomics beyond food analysis. TrAC Trends Anal. Chem. 2014, 59, 93–102. [Google Scholar] [CrossRef]
- Zhu, C.; Faillace, V.; Laus, F.; Bazzano, M.; Laghi, L. Characterization of trotter horses urine metabolome by means of proton nuclear magnetic resonance spectroscopy. Metabolomics 2018, 14, 106. [Google Scholar] [CrossRef]
- Zampiga, M.; Laghi, L.; Petracci, M.; Zhu, C.; Meluzzi, A.; Dridi, S.; Sirri, F. Effect of dietary arginine to lysine ratios on productive performance, meat quality, plasma and muscle metabolomics profile in fast-growing broiler chickens. J. Anim. Sci. Biotechnol. 2018, 9, 79. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Laghi, L.; Zhang, Z.; He, Y.; Wu, D.; Zhang, H.; Huang, Y.; Li, C.; Zou, L. First Steps toward the Giant Panda Metabolome Database: Untargeted Metabolomics of Feces, Urine, Serum, and Saliva by 1H NMR. J. Proteome Res. 2020, 19, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Bazzano, M.; Laghi, L.; Zhu, C.; Magi, G.E.; Serri, E.; Spaterna, A.; Tesei, B.; Laus, F. Metabolomics of tracheal wash samples and exhaled breath condensates in healthy horses and horses affected by equine asthma. J. Breath Res. 2018, 12, 046015. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Poulsen, N.A.; Larsen, L.B.; Bertram, H.C. Nuclear magnetic resonance metabonomics reveals strong association between milk metabolites and somatic cell count in bovine milk. J. Dairy Sci. 2013, 96, 290–299. [Google Scholar] [CrossRef]
- Xi, X.; Kwok, L.Y.; Wang, Y.; Ma, C.; Mi, Z.; Zhang, H. Ultra-performance liquid chromatography-quadrupole-time of flight mass spectrometry MSE-based untargeted milk metabolomics in dairy cows with subclinical or clinical mastitis. J. Dairy Sci. 2017, 100, 4884–4896. [Google Scholar] [CrossRef] [PubMed]
- Zandkarimi, F.; Vanegas, J.; Fern, X.; Maier, C.S.; Bobe, G. Metabotypes with elevated protein and lipid catabolism and inflammation precede clinical mastitis in prepartal transition dairy cows. J. Dairy Sci. 2018, 101, 5531–5548. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Li, C.; Wang, Y.; Laghi, L. Characterization of Yak Common Biofluids Metabolome by Means of Proton Nuclear Magnetic Resonance Spectroscopy. Metabolites 2019, 9, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kneen, M.A.; Annegarn, H.J. Algorithm for fitting XRF, SEM and PIXE X-ray spectra backgrounds. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 1996, 109–110, 209–213. [Google Scholar] [CrossRef]
- Liland, K.H.; Almøy, T.; Mevik, B.H. Optimal Choice of Baseline Correction for Multivariate Calibration of Spectra. Appl. Spectrosc. 2010, 64, 1007–1016. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application in1H NMR Metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2011; Volume 1, ISBN 3900051070. [Google Scholar]
- Box, G.E.P.; Cox, D.R. An Analysis of Transformations. J. R. Stat. Soc. Ser. B 2018, 26, 211–243. [Google Scholar] [CrossRef]
- Hubert, M.; Rousseeuw, P.J.; Vanden Branden, K. ROBPCA: A New Approach to Robust Principal Component Analysis. Technometrics 2005, 47, 64–79. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Jiang, H.; He, Q.; Wang, M.; Xue, J.; Liu, H.; Shi, K.; Wei, M.; Liang, S.; Zhang, L. Liquid Chromatography/Mass Spectrometry Reveals the Effect of Lactobacillus Treatment on the Faecal Metabolite Profile of Rats with Chronic Renal Failure. Nephron 2017, 135, 156–166. [Google Scholar] [CrossRef]
- Wang, Y.; Nan, X.; Zhao, Y.; Wang, H.; Wang, M.; Jiang, L.; Zhang, F.; Xue, F.; Hua, D.; Li, K.; et al. Coupling 16S rDNA Sequencing and Untargeted Mass Spectrometry for Milk Microbial Composition and Metabolites from Dairy Cows with Clinical and Subclinical Mastitis. J. Agric. Food Chem. 2020, 68, 8496–8508. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, V.D.; Ahir, V.B.; Koringa, P.G.; Jakhesara, S.J.; Rank, D.N.; Nauriyal, D.S.; Kunjadia, A.P.; Joshi, C.G. Milk microbiome signatures of subclinical mastitis-affected cattle analysed by shotgun sequencing. J. Appl. Microbiol. 2012, 112, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Zhang, H.; Zhang, Y.; Xiong, B.; Jiang, L. Microbiome and Metabolome Analyses of Milk from Dairy Cows with Subclinical Streptococcus agalactiae Mastitis—Potential Biomarkers. Front. Microbiol. 2019, 10, 2547. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.S.; Almstetter, M.F.; Schlamberger, G.; Nürnberger, N.; Dettmer, K.; Oefner, P.J.; Meyer, H.H.D.; Wiedemann, S.; Gronwald, W. Nuclear magnetic resonance and mass spectrometry-based milk metabolomics in dairy cows during early and late lactation. J. Dairy Sci. 2010, 93, 1539–1550. [Google Scholar] [CrossRef]
- Thomas, F.C.; Mudaliar, M.; Tassi, R.; McNeilly, T.N.; Burchmore, R.; Burgess, K.; Herzyk, P.; Zadoks, R.N.; Eckersall, P.D. Mastitomics, the integrated omics of bovine milk in an experimental model of: Streptococcus uberis mastitis: 3. Untargeted metabolomics. Mol. Biosyst. 2016, 12, 2762–2769. [Google Scholar] [CrossRef]
- Mansor, R. Proteomic and Metabolomic Studies on Milk during Bovine Mastitis. Ph.D. Thesis, University of Glasgow, Glasgow, UK, 2012. [Google Scholar]
- Ritter, W.; Hanni, H. The application of gas chromato-graphy in dairying. II. Detection and determination of volatile fatty acids in dairy products and cultures. Milchwissenschaft 1960, 15, 296–302. [Google Scholar]
- Brown, B. Basic Concepts in Biochemistry: A Student’s Survival Guide. Biochem. Educ. 1992, 20, 186. [Google Scholar] [CrossRef]
- Waisbren, S.E.; Noel, K.; Fahrbach, K.; Cella, C.; Frame, D.; Dorenbaum, A.; Levy, H. Phenylalanine blood levels and clinical outcomes in phenylketonuria: A systematic literature review and meta-analysis. Mol. Genet. Metab. 2007, 92, 63–70. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [Green Version]
- Keen, P.M.; Waterman, A.E.; Bourne, F.J. Oxygen concentration in milk of healthy and mastitic cows and implications of low oxygen tension for the killing of Staphylococcus aureus by bovine neutrophils. J. Dairy Res. 1988, 55, 513–519. [Google Scholar]
- Davis, S.R.; Farr, V.C.; Prosser, C.G.; Nicholas, G.D.; Turner, S.A.; Lee, J.; Hart, A.L. Milk L-lactate concentration is increased during mastitis. J. Dairy Res. 2004, 71, 175–181. [Google Scholar] [CrossRef]
- Thomas, F.C. Acute Phase Proteins, Proteomics and Metabolomics in the Diagnosis of Bovine Mastitis. Ph.D. Thesis, University of Glasgow, Glasgow, UK, 2015. [Google Scholar]
- Boudonck, K.J.; Mitchell, M.W.; Wulff, J.; Ryals, J.A. Characterization of the biochemical variability of bovine milk using metabolomics. Metabolomics 2009, 5, 375–386. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Frederiksen, P.D.; Clausen, M.R.; Larsen, L.B.; Bertram, H.C. Relationship between the Metabolite Profile and Technological Properties of Bovine Milk from Two Dairy Breeds Elucidated by NMR-Based Metabolomics. J. Agric. Food Chem. 2011, 59, 7360–7367. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Q.; Yin, P.; Xing, W.; Wu, Z.; Chen, S.; Lü, X.; Zhang, Y.; Lin, X.; Xu, G. Serum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseases. Anal. Bioanal. Chem. 2012, 403, 203–213. [Google Scholar] [CrossRef]
- Poortmans, J.R.; Kumps, A.; Duez, P.; Fofonka, A.; Carpentier, A.; Francaux, M. Effect of Oral Creatine Supplementation on Urinary Methylamine, Formaldehyde, and Formate. Med. Sci. Sports Exerc. 2005, 37, 1717–1720. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, Y.; Xia, C.; Zhang, H.; Qian, W.; Cao, Y. Pathway analysis of plasma different metabolites for dairy cow ketosis. Ital. J. Anim. Sci. 2016, 15, 545–551. [Google Scholar] [CrossRef] [Green Version]
- Graf, G.C.; Petersen, W.E. Changes in Respiration and Heart Rates, Body Temperatures, Plasma Lactic Acid Levels and Plasma Creatinine Levels Caused by Stress in Dairy Cattle. J. Dairy Sci. 1953, 36, 1036–1048. [Google Scholar] [CrossRef]
- Shennan, D.B.; Peaker, M. Transport of Milk Constituents by the Mammary Gland. Physiol. Rev. 2000, 80, 925–951. [Google Scholar] [CrossRef] [PubMed]
- Bannerman, D.D. Pathogen-dependent induction of cytokines and other soluble inflammatory mediators during intramammary infection of dairy cows. J. Anim. Sci. 2009, 87, 10–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Healthy | Clinical Mastitis | p Value | Trend | |
|---|---|---|---|---|
| Amino Acids, Peptides and Analogues | ||||
| Alanine | 5.31 × 10−2 (1.23 × 10−2) | 1.68 × 10−1 (1.04 × 10−1) | 0.2411 | ↑ |
| Arginine | 2.30 × 10−2 (4.72 × 10−3) | 2.04 × 10−1 (1.13 × 10−1) | 0.0190 | ↑ |
| Betaine | 4.64 × 10−2 (8.52 × 10−4) | 4.57 × 10−2 (4.06 × 10−2) | 0.9821 | ↓ |
| Creatine | 8.35 × 10−1 (7.90 × 10−2) | 2.06 × 10−1 (7.82 × 10−2) | 0.0106 | ↓ |
| Creatine phosphate | 1.13 × 10−1 (7.85 × 10−2) | 4.49 × 10−2 (2.36 × 10−2) | 0.0589 | ↓ |
| Creatinine | 1.12 × 10−1 (1.44 × 10−2) | 2.39 × 10−2 (1.45 × 10−2) | 0.0000 | ↓ |
| Glutamate | 7.46 × 10−1 (3.93 × 10−1) | 6.57 × 10−1 (6.59 × 10−1) | 0.6295 | ↓ |
| Glycine | 1.63 (1.37) | 2.65 (2.49) | 0.0000 | ↑ |
| Histidine | 9.35 × 10−2 (1.60 × 10−2) | 2.88 × 10−2 (5.00 × 10−2) | 0.0278 | ↓ |
| Isoleucine | 4.54 × 10−3 (3.99 × 10−3) | 8.45 × 10−2 (7.13 × 10−2) | 0.0021 | ↑ |
| Leucine | 1.10 × 10−1 (2.69 × 10−2) | 5.27 × 10−1 (3.93 × 10−2) | 0.0003 | ↑ |
| Lysine | 1.58 × 10−1 (1.61 × 10−2) | 2.68 × 10−1 (1.86 × 10−1) | 0.1177 | ↑ |
| N,N-Dimethylglycine | 3.16 × 10−3 (1.42 × 10−4) | 3.06 × 10−3 (1.40 × 10−2) | 0.9092 | ↓ |
| Phenylalanine | 2.14 × 10−2 (7.10 × 10−3) | 6.47 × 10−2 (6.65 × 10−2) | 0.2731 | ↑ |
| Proline | 2.46 × 10−3 (2.21 × 10−3) | 8.15 × 10−2 (8.95 × 10−2) | 0.0028 | ↑ |
| Tyrosine | 3.90 × 102 (1.48 × 10−3) | 8.57 × 10−2 (4.20 × 10−2) | 0.0214 | ↑ |
| Valine | 1.77 × 10−2 (6.67 × 10−3) | 1.46 × 10−1 (1.18 × 10−1) | 0.0017 | ↑ |
| Carbohydrates and Derivates | ||||
| Acetone | 9.82 × 10−3 (1.27 × 10−3) | 4.81 × 10−3 (6.18 × 10−3) | 0.0728 | ↓ |
| Galactose | 7.32 × 10−1 (1.91 × 10−1) | 2.71 × 10−1 (3.71 × 10−1) | 0.3737 | ↓ |
| Glucose | 6.77 × 10−1 (2.96 × 10−2) | 1.88 × 10−1 (2.53 × 10−1) | 0.1906 | ↓ |
| Glucose-1-phosphate | 1.58 × 10−1 (7.73 × 10−2) | 1.05 × 10−2 (7.54 × 10−3) | 0.0675 | ↓ |
| Lactose | 119 (8.77) | 15.8 (23.3) | 0.0012 | ↓ |
| Maltose | 1.25 × 10−1 (7.10 × 10−2) | 4.08 × 10−3 (3.78 × 10−3) | 0.0121 | ↓ |
| Xylose | 1.73 (4.06 × 10−1) | 5.43 × 10−1 (4.60 × 10−1) | 0.1879 | ↓ |
| Organic Acids and Derivates | ||||
| 2-Hydroxyisobutyrate | 3.34 × 10−3 (8.25 × 10−4) | 2.12 × 10−2 (8.16 × 10−1) | 0.2667 | ↑ |
| 2-Oxoglutarate | 2.46 × 10−1 (6.12 × 10−2) | 3.46 × 10−2 (1.82 × 10−2) | 0.0000 | ↓ |
| 3-Hydroxybutyrate | 9.40 × 10−2 (2.11 × 10−2) | 1.83 × 10−1 (1.14 × 10−1) | 0.0012 | ↑ |
| Acetate | 3.68 × 10−2 (9.56 × 10−3) | 3.91 × 10−1 (4.18 × 10−1) | 0.1093 | ↑ |
| Citrate | 8.92 (1.32) | 1.63 (1.02) | 0.0002 | ↓ |
| Formate | 3.19 × 10−2 (5.96 × 10−3) | 6.40 × 10−2 (2.47 × 10−1) | 0.4165 | ↑ |
| Fumarate | 2.8910−2 (3.40 × 10−4) | 6.9710−3 (4.40 × 10−3) | 0.0002 | ↑ |
| Hippurate | 5.76 × 10−2 (3.27 × 10−2) | 4.03 × 10−2 (3.78 × 10−2) | 0.0517 | ↓ |
| Iso-butyrate | 2.57 × 10−3 (8.42 × 10−4) | 5.77 × 10−3 (3.84 × 10−3) | 0.1710 | ↑ |
| Lactate | 7.84 × 10−2 (2.50 × 10−2) | 3.10 (5.14) | 0.0129 | ↑ |
| Malonate | 1.35 × 10−2 (1.22 × 10−2) | 5.62 × 10−3 (3.09 × 10−3) | 0.0644 | ↓ |
| Pimelate | 1.56 × 10−1 (2.48 × 10−3) | 4.80 × 10−2 (2.35 × 10−2) | 0.1168 | ↓ |
| Propionate | 2.01 × 10−3 (3.68 × 10−4) | 4.35 × 10−4 (1.08 × 10−4) | 0.1625 | ↓ |
| Pyruvate | 2.01 × 10−2 (4.79 × 10−3) | 3.09 × 10−2 (1.39 × 10−2) | 0.0128 | ↑ |
| Succinate | 2.97 × 10−2 (4.64 × 10−3) | 3.26 × 10−2 (1.13 × 10−1) | 0.6861 | ↑ |
| Nucleosides, Nucleotides and Analogues | ||||
| Cytidine | 2.95 × 10−2 (1.54 × 10−3) | 9.68 × 10−3 (1.51 × 10−3) | 0.0005 | ↓ |
| Dimethylamine | 2.42 × 10−3 (4.04 × 10−4) | 6.86 × 10−3 (9.24 × 10−3) | 0.0414 | ↑ |
| N-Acetylglucosamine | 9.22 × 10−1 (4.77 × 10−1) | 4.97 × 10−1 (3.64 × 10−1) | 0.0311 | ↓ |
| Trimethylamine-N-oxide | 1.69 × 10−1 (6.86 × 10−3) | 4.96 × 10−2 (2.54 × 10−2) | 0.0002 | ↓ |
| Uridine | 2.94 × 10−2 (1.35 × 10−2) | 2.92 × 10−2 (4.66 × 10−1) | 0.6721 | ↓ |
| Miscellaneous | ||||
| Carnitine | 2.60 × 10−1 (1.29 × 10−1) | 1.43 × 10−2 (2.21 × 10−2) | 0.0485 | ↓ |
| Choline | 3.37 × 10−1 (3.82 × 10−2) | 7.41 × 10−2 (3.43 × 10−2) | 0.0009 | ↑ |
| cis-Aconitate | 1.52 × 10−1 (1.69 × 10−2) | 3.46 × 10−2 (2.18 × 10−2) | 0.0000 | ↓ |
| Dimethyl sulfone | 1.73 × 10−2 (9.08 × 10−3) | 7.56 × 10−3 (4.14 × 10−3) | 0.0035 | ↓ |
| Ethanol | 4.37 × 10−2 (3.45 × 10−3) | 7.59 × 10−2 (8.97 × 10−2) | 0.5625 | ↑ |
| O-Acetyl-carnitine | 5.54 × 10−2 (6.16 × 10−3) | 1.48 × 10−2 (1.14 × 10−2) | 0.0451 | ↓ |
| Propylene glycol | 1.03 × 10−2 (2.09 × 10−3) | 2.76 × 10−2 (4.99 × 10−1) | 0.1390 | ↑ |
| trans-Aconitate | 2.31 × 10−2 (1.97 × 10−3) | 1.25 × 10−2 (6.68 × 10−3) | 0.1530 | ↓ |
| sn-Glycero-3-phosphocholine | 1.23 (1.48 × 10−1) | 2.30 × 10−1 (1.20 × 10−1) | 0.0799 | ↓ |
| Uricil | 1.35 × 10−2 (2.23 × 10−3) | 1.60 × 10−2 (1.49 × 10−2) | 0.7987 | ↑ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.; Tang, K.; Lu, X.; Tang, J.; Laghi, L. An Untargeted Metabolomics Investigation of Milk from Dairy Cows with Clinical Mastitis by 1H-NMR. Foods 2021, 10, 1707. https://doi.org/10.3390/foods10081707
Zhu C, Tang K, Lu X, Tang J, Laghi L. An Untargeted Metabolomics Investigation of Milk from Dairy Cows with Clinical Mastitis by 1H-NMR. Foods. 2021; 10(8):1707. https://doi.org/10.3390/foods10081707
Chicago/Turabian StyleZhu, Chenglin, Kaiwei Tang, Xuan Lu, Junni Tang, and Luca Laghi. 2021. "An Untargeted Metabolomics Investigation of Milk from Dairy Cows with Clinical Mastitis by 1H-NMR" Foods 10, no. 8: 1707. https://doi.org/10.3390/foods10081707
APA StyleZhu, C., Tang, K., Lu, X., Tang, J., & Laghi, L. (2021). An Untargeted Metabolomics Investigation of Milk from Dairy Cows with Clinical Mastitis by 1H-NMR. Foods, 10(8), 1707. https://doi.org/10.3390/foods10081707