Evaluation of PhageDX Salmonella Assay for Salmonella Detection in Hydroponic Curly Lettuce
Abstract
:1. Introduction
2. Materials and Methods
2.1. PhageDx Salmonella Assay
2.2. In Vitro Assay for Determination of Detection Limit
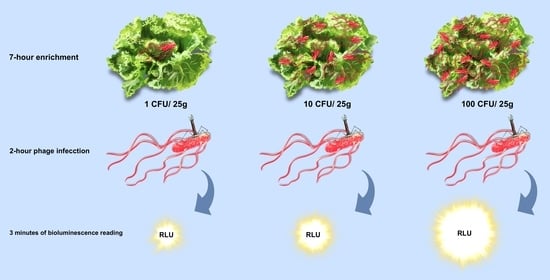
2.3. Salmonella Detection on Hydroponic Curly Lettuce
2.4. Bioluminescence Assay
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hassenberg, K.; Praeger, U.; Herppich, W.B. Effect of chlorine dioxide treatment on human pathogens on iceberg lettuce. Foods 2021, 10, 574. [Google Scholar] [CrossRef]
- Mir, S.A.; Shah, M.A.; Mir, M.M. Microgreens: Production, shelf life and bioactive components. Crit. Rev. Food Sci. Nutr. 2017, 57, 2730–2736. [Google Scholar] [CrossRef]
- Rico, D.; Martin-Diana, A.B.; Barat, J.; Barry-Ryan, C. Extending and measuring the quality of fresh-cut fruit and vegetables: A review. Trends Food Sci. Technol. 2007, 18, 373–386. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, M.W.; Chakraborty, I.; Ayala-Zavala, J.F.; Dhua, R.S. Advances in minimal processing of fruits and vegetables: A review. J. Sci. Ind. Res. 2011, 70, 823–834. [Google Scholar]
- Olaimat, A.N.; Holley, R.A. Factors influencing the microbial safety of fresh produce: A review. Food Microbiol. 2012, 32, 1–19. [Google Scholar] [CrossRef]
- Elias, S.O.; Noronha, T.B.; Tondo, E.C. Risk of infection with Salmonella and Escherichia coli O157: H7 due to consumption of lettuce in southern Brazil. MOJ Food Process. Technol. 2021, 9, 57–66. [Google Scholar] [CrossRef]
- Machado-Moreira, B.; Richards, K.; Brennan, F.; Abram, F.; Burgess, C.M. Microbial contamination of fresh produce: What, where, and how? Compr. Rev. Food Sci. Food Saf. 2019, 18, 1727–1750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mafi, N.; Orenstein, R. Salmonellosis. In Encyclopedia of Gastroenterology, 2nd ed.; Kuipers, E.J., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 384–391. [Google Scholar]
- Andrews, W.H.; Wang, H.; Jacobson, A.; Hammack, T. BAM Chapter 5: Salmonella. In Bacteriological Analytical Manual; U.S. Food & Drug Administration: Silver Spring, MD, USA, 2018. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-5-salmonella (accessed on 11 May 2021).
- Valderrama, W.B.; Dudley, E.G.; Doores, S.; Cutter, C.N. Commercially available rapid methods for detection of selected foodborne pathogens. Crit. Rev. Food Sci. Nutr. 2016, 56, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, L.; Bolton, D.; McAuliffe, O.; Coffey, A. Bacteriophages in food applications: From foe to friend. Annu. Rev. Food Sci. Technol. 2019, 10, 151–172. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.M.; Gil, J.; Brown, M.; Tondo, E.C.; de Aquino, N.S.M.; Eisenberg, M.; Erickson, S. Accurate and sensitive detection of Salmonella in foods by engineered bacteriophages. Sci. Rep. 2020, 10, 17463. [Google Scholar] [CrossRef]
- APHA—American Public Health Association. Compendium of Methods for Microbiological Examination of Food, 3rd ed.; Vanderzant, C., Splittstoesser, D.F., Eds.; American Public Health Association: Washington, DC, USA, 1992. [Google Scholar]
- Heller, K.J. Molecular interaction between bacteriophage and the gram-negative cell envelope. Arch. Microbiol. 1992, 158, 235–248. [Google Scholar] [CrossRef]
- Silva, J.B.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [Green Version]
- Hyman, P.; Abedon, S.T. Chapter 7: Bacteriophage host range and bacterial resistance. In Advances in Applied Microbiology; Laskin, A.I., Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2010; Volume 70, pp. 217–248. [Google Scholar] [CrossRef]
- Mostafidi, M.; Sanjabi, M.R.; Shirkhan, F.; Zahedi, M.T. A review of recent trends in the development of the microbial safety of fruits and vegetables. Trends Food Sci. Technol. 2020, 103, 321–332. [Google Scholar] [CrossRef]
- Hohnadel, M.; Maumy, M.; Chollet, R. Development of a micromanipulation method for single cell isolation of prokaryotes and its application in food safety. PLoS ONE 2018, 13, e0198208. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, B.M.; de Castro Antunes Marques Fernandez, E.; Fernandez, A.T. Avaliação da eficiência antibacteriana de fermentados acéticos comerciais em saladas de alface (Lactuca sativa) comercializadas na cidade de Duque de Caxias, Rio de Janeiro. Vigil. Sanit. Debate Soc. Ciênc. Tecnol. (Health Surveill. Debate Soc. Sci. Technol.) Visa Debate 2019, 7, 53–59. [Google Scholar] [CrossRef]
- Schuh, V.; Schuh, J.; Fronza, N.; Foralosso, F.B.; Verruck, S.; Vargas Junior, A.; da Silveira, S.M. Evaluation of the microbiological quality of minimally processed vegetables. Food Sci. Technol. 2020, 40, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Maffei, D.F.; de Arruda Silveira, N.F.; Catanozi, M.d.P.L.M. Microbiological quality of organic and conventional vegetables sold in Brazil. Food Control 2013, 29, 226–230. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Maestu, A.; Fuciños, P.; Azinheiro, S.; Carvalho, C.; Carvalho, J.; Prado, M. Specific detection of viable Salmonella Enteritidis by phage amplification combined with qPCR (PAA-qPCR) in spiked chicken meat sample. Food Control 2019, 99, 79–83. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yan, C.; Yang, H.; Yu, J.; Wei, H. Rapid and selective detection of E. coli O157:H7 combining phagomagnetic separation with enzymatic colorimetry. Food Chem. 2017, 234, 332–338. [Google Scholar] [CrossRef]
- Rees, J.C.; Voorhees, K.J. Simultaneous detection of two bacterial pathogens using bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 2757–2761. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, M.; Kim, S.; Ryu, S. Sensitive detection of viable Escherichia coli O157:H7 from foods using a luciferase-reporter phage phiV10lux. Int. J. Food Microbiol. 2017, 254, 11–17. [Google Scholar] [CrossRef] [PubMed]
| Sample | Inoculum | Number of Replicates | Avg. RLU | SD | % CV |
|---|---|---|---|---|---|
| S. Minnesota | Low | 30 | 187 | 41 | 22 |
| Medium | 30 | 223 | 89 | 40 | |
| High | 12 | 1475 | 953 | 65 | |
| S. Enteritidis | Low | 30 | 182 | 45 | 25 |
| Medium | 30 | 246 | 168 | 68 | |
| High | 12 | 724 | 339 | 47 | |
| S. Saintpaul | Low | 30 | 198 | 115 | 58 |
| Medium | 30 | 407 | 398 | 98 | |
| High | 12 | 3388 | 2061 | 61 | |
| S. Infantis | Low | 30 | 214 | 169 | 79 |
| Medium | 30 | 479 | 415 | 87 | |
| High | 12 | 1914 | 1324 | 69 | |
| S. Heidelberg | Low | 30 | 325 | 220 | 68 |
| Medium | 30 | 873 | 426 | 49 | |
| High | 12 | 6158 | 1729 | 28 | |
| S. Typhimurium | Low | 30 | 1814 | 1305 | 72 |
| Medium | 30 | 14,648 | 9257 | 63 | |
| High | 12 | 16,715 | 15,1623 | 907 | |
| Salmonella cocktail | Low | 30 | 2323 | 339 | 15 |
| Medium | 30 | 3054 | 1559 | 51 | |
| High | 12 | 26,543 | 9980 | 38 | |
| Lettuce | Low | 30 | 1914 | 6813 | 356 |
| Medium | 30 | 7347 | 6207 | 84 | |
| High | 12 | 106,579 | 68,315 | 64 |
| Samples | Negative Control | Low | Medium | High |
|---|---|---|---|---|
| S. Minnesota | 0 (0/2) | 0 (0/30) | 0 (0/30) | 75 (9/12) |
| S. Enteritidis | 0 (0/2) | 0 (0/30) | 6.7 (2/30) | 50 (6/12) |
| S. Saintpaul | 0 (0/2) | 3.0 (1/30) | 16.7 (5/30) | 100 (12/12) |
| S. Infantis | 0 (0/2) | 3.0 (1/30) | 16.7 (5/30) | 100 (12/12) |
| S. Heidelberg | 0 (0/2) | 10 (3/30) | 60 (18/30) | 100 (12/12) |
| S. Typhimurium | 0 (0/2) | 83.3 (25/30) | 100 (12/12) | 100 (12/12) |
| Salmonella cocktail | 0 (0/2) | 63 (19/30) | 93 (28/30) | 100 (12/12) |
| Lettuces | 0 (0/2) | 30 (9/30) | 100 (12/12) | 100 (12/12) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Aquino, N.S.M.; Elias, S.d.O.; Tondo, E.C. Evaluation of PhageDX Salmonella Assay for Salmonella Detection in Hydroponic Curly Lettuce. Foods 2021, 10, 1795. https://doi.org/10.3390/foods10081795
de Aquino NSM, Elias SdO, Tondo EC. Evaluation of PhageDX Salmonella Assay for Salmonella Detection in Hydroponic Curly Lettuce. Foods. 2021; 10(8):1795. https://doi.org/10.3390/foods10081795
Chicago/Turabian Stylede Aquino, Nathanyelle Soraya Martins, Susana de Oliveira Elias, and Eduardo Cesar Tondo. 2021. "Evaluation of PhageDX Salmonella Assay for Salmonella Detection in Hydroponic Curly Lettuce" Foods 10, no. 8: 1795. https://doi.org/10.3390/foods10081795
APA Stylede Aquino, N. S. M., Elias, S. d. O., & Tondo, E. C. (2021). Evaluation of PhageDX Salmonella Assay for Salmonella Detection in Hydroponic Curly Lettuce. Foods, 10(8), 1795. https://doi.org/10.3390/foods10081795