Widely Targeted Lipidomics and Transcriptomics Analysis Revealed Changes of Lipid Metabolism in Spleen Dendritic Cells in Shrimp Allergy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
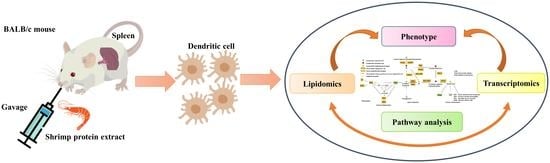
2.2. Experimental Scheme
2.3. sIgE and sIgG1 Analysis
2.4. Cytokine Measurement
2.5. Assessment of Clinical Anaphylaxis and Body Temperature
2.6. Preparation of Single Cell Suspension of Spleen
2.7. Fluorescent Staining for Flow Cytometer Analysis
2.8. Isolation of Spleen DCs by Fluorescene-Activated Cell Sorting
2.9. Lipid Extraction and Widely Targeted Lipidomics Analysis
2.10. RNA Sequence Analysis
2.11. Correlation Analysis of Transcriptome and Lipidome
2.12. Dendritic Cell Line DC2.4 Culture, Cell Treatment, and RNA Extraction
2.13. RT-qPCR
2.14. Statistics
3. Results
3.1. Shrimp Allergic Mouse Model Showed Typical Allergic Reactions
3.2. SA Affected Lipid Metabolism and Immune Function of Spleen DCs
3.3. Lipid Profile Distinguished Spleen DCs in SA from Normal
3.4. Functional Enrichment of DEGs in SA
3.5. GSEA of B220, CD40, and CD68 Related Gene Sets
3.6. Correlation Analysis Based on Transcriptome and Lipidome
3.7. Pathway Analysis Based on Lipidome and Transcriptome
3.8. Glyceryl Trioleate and C16 Ceramide Affected the Immune Function of Dendritic Cell Line DC2.4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pawankar, R.; Canonica, G.; Holgate, S.; Lockey, R.F.; Blaiss, M. World Allergy Organization (WAO) White Book on Allergy, Update; World Allergy Organization: Milwaukee, WI, USA, 2013. [Google Scholar]
- WHO; FAO. Joint FAO/WHO Expert Consultation on Allergenicity of Foods Derived from Biotechnology. In Proceedings of the Evaluation of Allergenicity of Genetically Modified Foods: Report of a Joint FAO/WHO Expert Consultation on Allergenicity of Foods Derived from Biotechnology, Rome, Italy, 22–25 January 2001. [Google Scholar]
- Cardona, V.; Ansotegui, I.J.; Ebisawa, M.; El-Gamal, Y.; Rivas, M.F.; Fineman, S.; Geller, M.; Gonzalez-Estrada, A.; Greenberger, P.A.; Borges, M.S. World allergy organization anaphylaxis guidance 2020. World Allergy Organ. J. 2020, 13, 100472. [Google Scholar] [CrossRef] [PubMed]
- Sicherer, S.H.; Muñoz-Furlong, A.; Sampson, H.A. Prevalence of seafood allergy in the United States determined by a random telephone survey. J. Allergy Clin. Immunol. 2004, 114, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Ruethers, T.; Taki, A.C.; Johnston, E.B.; Nugraha, R.; Le, T.T.K.; Kalic, T.; McLean, T.R.; Kamath, S.D.; Lopata, A.L. Seafood allergy: A comprehensive review of fish and shellfish allergens. Mol. Immunol. 2018, 100, 28–57. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.; Tham, E.H.; Lee, B.W. An update on shellfish allergy. Curr. Opin. Allergy Clin. Immunol. 2019, 19, 236–242. [Google Scholar] [CrossRef]
- Sena-Torralba, A.; Pallás-Tamarit, Y.; Morais, S.; Maquieira, Á. Recent advances and challenges in food-borne allergen detection. TrAC Trends Anal. Chem. 2020, 132, 116050. [Google Scholar] [CrossRef]
- Amin, K. The role of mast cells in allergic inflammation. Respir. Med. 2012, 106, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, J.; Nguyen, A.H.; Rehman, A.; Ochi, A.; Jamal, M.; Graffeo, C.S.; Henning, J.R.; Zambirinis, C.P.; Fallon, N.C.; Barilla, R.; et al. Dendritic cell populations with different concentrations of lipid regulate tolerance and immunity in mouse and human liver. Gastroenterology 2012, 143, 1061–1072. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Gu, Y.; Wang, J.; Chen, C.; Han, S.; Che, H. Effects of Fatty Acid Oxidation and Its Regulation on Dendritic Cell-Mediated Immune Responses in Allergies: An Immunometabolism Perspective. J. Immunol. Res. 2021, 2021, 7483865. [Google Scholar] [CrossRef]
- Zhang, C.; Yue, C.; Herrmann, A.; Song, J.; Egelston, C.; Wang, T.; Zhang, Z.; Li, W.; Lee, H.; Aftabizadeh, M. STAT3 activation-induced fatty acid oxidation in CD8+ T effector cells is critical for obesity-promoted breast tumor growth. Cell Metab. 2020, 31, 148–161. [Google Scholar] [CrossRef]
- Lee, J.; Choi, J.; Alpergin, E.S.S.; Zhao, L.; Hartung, T.; Scafidi, S.; Riddle, R.C.; Wolfgang, M.J. Loss of hepatic mitochondrial long-chain fatty acid oxidation confers resistance to diet-induced obesity and glucose intolerance. Cell Rep. 2017, 20, 655–667. [Google Scholar] [CrossRef] [Green Version]
- Arita, M. Eosinophil polyunsaturated fatty acid metabolism and its potential control of inflammation and allergy. Allergol. Int. 2016, 65, 2–5. [Google Scholar] [CrossRef] [Green Version]
- Herber, D.L.; Cao, W.; Nefedova, Y.; Novitskiy, S.V.; Nagaraj, S.; Tyurin, V.A.; Corzo, A.; Cho, H.-I.; Celis, E.; Lennox, B. Lipid accumulation and dendritic cell dysfunction in cancer. Nat. Med. 2010, 16, 880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, E.; Zhang, G.; Huang, J.; Yang, X.; Peng, L.; Huang, X.; Luo, X.; Ren, J.; Huang, R.; Yang, L. Immunomodulatory effect of oleoylethanolamide in dendritic cells via TRPV1/AMPK activation. J. Cell. Physiol. 2019, 234, 18392–18407. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Xiao, C.; Evans, K.S.; Theivanthiran, T.; DeVito, N.; Holtzhausen, A.; Liu, J.; Liu, X.; Boczkowski, D.; Nair, S.; et al. Paracrine Wnt5a-β-Catenin Signaling Triggers a Metabolic Program that Drives Dendritic Cell Tolerization. Immunity 2018, 48, 147–160.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammad, H.; de Heer, H.J.; Soullié, T.; Angeli, V.; Trottein, F.; Hoogsteden, H.C.; Lambrecht, B.N. Activation of peroxisome proliferator-activated receptor-γ in dendritic cells inhibits the development of eosinophilic airway inflammation in a mouse model of asthma. Am. J. Pathol. 2004, 164, 263–271. [Google Scholar] [CrossRef]
- Jiang, S.; Han, S.; Chen, J.; Li, X.; Che, H. Inhibition effect of blunting Notch signaling on food allergy through improving TH1/TH2 balance in mice. Ann. Allergy, Asthma Immunol. 2017, 118, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Jiang, S.; Wang, J.; Chen, C.; Han, S.; Che, H. Cholera toxin induces food allergy through Th2 cell differentiation which is unaffected by Jagged2. Life Sci. 2020, 263, 118514. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Pan, T.; Cao, M.; Liu, Q.; Zhang, L.; Liu, G. Suppression of Th2 immune responses by the sulfated polysaccharide from Porphyra haitanensis in tropomyosin-sensitized mice. Int. Immunopharmacol. 2015, 24, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Schofield, B.H.; Huang, C.-K.; Kleiner, G.I.; Sampson, H.A. A murine model of IgE-mediated cow’s milk hypersensitivity. J. Allergy Clin. Immunol. 1999, 103, 206–214. [Google Scholar] [CrossRef]
- Tavernier, S.J.; Osorio, F.; Janssens, S.; Lambrecht, B.N. Isolation of splenic dendritic cells using fluorescence-activated cell sorting. Bio-Protoc. 2015, 5, e1415. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Hu, S.; Li, L.; Han, C.; Liu, H.; He, H.; Xia, L.; Hu, J.; Hu, B.; Ran, M. Lipidomics profiling of goose granulosa cell model of stearoyl-CoA desaturase function identifies a pattern of lipid droplets associated with follicle development. Cell Biosci. 2021, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Wang, K.; Lyu, S.; Ren, L.; Huang, C.; Pei, D.; Xing, Y.; Wang, Y.; Xu, Y. Comparison analysis of widely-targeted metabolomics revealed the variation of potential astringent ingredients and their dynamic accumulation in the seed coats of both Carya cathayensis and Carya illinoinensis. Food Chem. 2022, 374, 131688. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Han, B.; Li, X.; Ai, R.-S.; Deng, S.-Y.; Ye, Z.-Q.; Deng, X.; Ma, W.; Xiao, S.; Wang, J.-Z.; Wang, L.-M. Atmospheric particulate matter aggravates CNS demyelination through involvement of TLR-4/NF-kB signaling and microglial activation. Elife 2022, 11, e72247. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, M.R.; Sambrook, J. Purification of total RNA from mammalian cells and tissues. Cold Spring Harb. Protoc. 2020, 2020, pdb-prot101659. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.I.; Sindher, S.B.; Chinthrajah, R.S.; Nadeau, K.; Davis, C.M. Shrimp-allergic patients in a multi-food oral immunotherapy trial. Pediatr. Allergy Immunol. 2022, 33, e13679. [Google Scholar] [CrossRef]
- Hoffmann, K. Plant allergens and pathogenesis related proteins. Int. Arch. Allergy Immunol. 2000, 122, 155–166. [Google Scholar] [CrossRef]
- Sampson, H.A.; O’Mahony, L.; Burks, A.W.; Plaut, M.; Lack, G.; Akdis, C.A. Mechanisms of food allergy. J. Allergy Clin. Immunol. 2018, 141, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Long, F.; Yang, X.; Wang, R.; Hu, X.; Chen, F. Effects of combined high pressure and thermal treatments on the allergenic potential of shrimp (Litopenaeus vannamei) tropomyosin in a mouse model of allergy. Innov. Food Sci. Emerg. Technol. 2015, 29, 119–124. [Google Scholar] [CrossRef]
- Schiavi, E.; Barletta, B.; Butteroni, C.; Corinti, S.; Boirivant, M.; Di Felice, G. Oral therapeutic administration of a probiotic mixture suppresses established Th2 responses and systemic anaphylaxis in a murine model of food allergy. Allergy 2011, 66, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; van Nimwegen, M.; Willart, M.A.M.; Muskens, F.; Boon, L.; Smit, J.J.; Coyle, A.; Clausen, B.E.; Hoogsteden, H.C.; Lambrecht, B.N. An anti-inflammatory role for plasmacytoid dendritic cells in allergic airway inflammation. J. Immunol. 2009, 183, 1074–1082. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Zheng, X.; Zhang, X.; Zhang, Z.-X.; Ichim, T.E.; Sun, H.; Nakamura, Y.; Inagaki, A.; Beduhn, M.; Shunnar, A. A novel allergen-specific therapy for allergy using CD40-silenced dendritic cells. J. Allergy Clin. Immunol. 2010, 125, 737–743. [Google Scholar] [CrossRef]
- Bublin, M.; Eiwegger, T.; Breiteneder, H. Do lipids influence the allergic sensitization process? J. Allergy Clin. Immunol. 2014, 134, 521–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koplin, J.J.; Peters, R.L. Explaining the link between maternal lipid profiles and food allergy in offspring. J. Allergy Clin. Immunol. 2019, 144, 661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, X.; Liang, L.; Sun, Q.; Keet, C.A.; Tsai, H.-J.; Ji, Y.; Wang, G.; Ji, H.; Clish, C.; Pearson, C. Maternal triacylglycerol signature and risk of food allergy in offspring. J. Allergy Clin. Immunol. 2019, 144, 729–737. [Google Scholar] [CrossRef] [Green Version]
- Bishop, W.R.; Bell, R.M. Functions of diacylglycerol in glycerolipid metabolism, signal transduction and cellular transformation. Oncogene Res. 1988, 2, 205–218. [Google Scholar]
- Bao, R.; Hesser, L.A.; He, Z.; Zhou, X.; Nadeau, K.C.; Nagler, C.R. Fecal microbiome and metabolome differ in healthy and food-allergic twins. J. Clin. Investig. 2021, 131, e141935. [Google Scholar] [CrossRef]
- Winkler, S.C.; Shimobayashi, E.; Kapfhammer, J.P. PKCγ-Mediated Phosphorylation of CRMP2 Regulates Dendritic Outgrowth in Cerebellar Purkinje Cells. Mol. Neurobiol. 2020, 57, 5150–5166. [Google Scholar] [CrossRef]
- WANG, X.; CAO, Y. Research progress in the roles of miR-155 in atherosclerosis. J. Clin. Pathol. Res. 2015, 35, 2185–2190. [Google Scholar]
- Newton, A.C. Protein kinase C: Structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 2001, 101, 2353–2364. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, Z.; Kuang, P.; Shi, X.; Wang, Z.; Guo, L. Regulation of lipid metabolism in diabetic rats by Arctium lappa L. polysaccharide through the PKC/NF-κB pathway. Int. J. Biol. Macromol. 2019, 136, 115–122. [Google Scholar] [CrossRef] [PubMed]
- La Porta, C.A.; Comolli, R. PKC-dependent modulation of IkB alpha-NFkB pathway in low metastatic B16F1 murine melanoma cells and in highly metastatic BL6 cells. Anticancer Res. 1998, 18, 2591–2597. [Google Scholar] [PubMed]
- Kitatani, K.; Usui, T.; Sriraman, S.K.; Toyoshima, M.; Ishibashi, M.; Shigeta, S.; Nagase, S.; Sakamoto, M.; Ogiso, H.; Okazaki, T. Ceramide limits phosphatidylinositol-3-kinase C2β-controlled cell motility in ovarian cancer: Potential of ceramide as a metastasis-suppressor lipid. Oncogene 2016, 35, 2801–2812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Oh, J.E.; Kim, S.W.; Chun, Y.J.; Kim, M.Y. Ceramide induces p38 MAPK-dependent apoptosis and Bax translocation via inhibition of Akt in HL-60 cells. Cancer Lett. 2008, 260, 88–95. [Google Scholar] [CrossRef]
- Ozes, O.N.; Mayo, L.D.; Gustin, J.A.; Pfeffer, S.R.; Pfeffer, L.M.; Donner, D.B. NF-κB activation by tumour necrosis factor requires the Akt serine–threonine kinase. Nature 1999, 401, 82–85. [Google Scholar] [CrossRef]
- Hsu, S.-Y.; Lee, S.-C.; Liu, H.-C.; Peng, S.-F.; Chueh, F.-S.; Lu, T.-J.; Lee, H.-T.; Chou, Y.-C. Phenethyl Isothiocyanate Suppresses the Proinflammatory Cytokines in Human Glioblastoma Cells through the PI3K/Akt/NF-κB Signaling Pathway In Vitro. Oxid. Med. Cell. Longev. 2022, 2022, 1–11. [Google Scholar] [CrossRef]
- Shimizu, K.; Konno, S.; Ozaki, M.; Umezawa, K.; Yamashita, K.; Todo, S.; Nishimura, M. Dehydroxymethylepoxyquinomicin (DHMEQ), a novel NF-kappaB inhibitor, inhibits allergic inflammation and airway remodelling in murine models of asthma. Clin. Exp. Allergy 2012, 42, 1273–1281. [Google Scholar] [CrossRef]
- Ma, S.Q.; Wei, H.L.; Zhang, X. TLR2 regulates allergic airway inflammation through NF-kappaB and MAPK signaling pathways in asthmatic mice. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3138–3146. [Google Scholar]
- Liang, Y.; Zhou, Y.; Shen, P. NF-kappaB and its regulation on the immune system. Cell Mol Immunol 2004, 1, 343–350. [Google Scholar] [PubMed]
- Yu, Y.; Li, J.; Liu, C. Oxytocin suppresses epithelial cell-derived cytokines production and alleviates intestinal inflammation in food allergy. Biochem. Pharmacol. 2022, 195, 114867. [Google Scholar] [CrossRef] [PubMed]
- Abboushi, N.; El-Hed, A.; El-Assaad, W.; Kozhaya, L.; El-Sabban, M.E.; Bazarbachi, A.; Badreddine, R.; Bielawska, A.; Usta, J.; Dbaibo, G.S. Ceramide inhibits IL-2 production by preventing protein kinase C-dependent NF-κB activation: Possible role in protein kinase Cθ regulation. J. Immunol. 2004, 173, 3193–3200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, L.; Han, F.; Gong, L.; Lv, Y.; Wan, Z.; Liu, H.; Ren, L.; Yang, S.; Zhang, W.; Li, T. Ceramide induces MMP-9 expression through JAK2/STAT3 pathway in airway epithelium. Lipids Health Dis. 2020, 19, 1–9. [Google Scholar] [CrossRef]
- Wen, Z.; Darnell, J.E., Jr. Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res. 1997, 25, 2062–2067. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Herrmann, A.; Deng, J.-H.; Kujawski, M.; Niu, G.; Li, Z.; Forman, S.; Jove, R.; Pardoll, D.M.; Yu, H. Persistently activated Stat3 maintains constitutive NF-κB activity in tumors. Cancer Cell 2009, 15, 283–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2012, 41, D377–D386. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.K.; Kammer, G.M.; Dennis, G.; Tsokos, G.C. Abnormal NF-κB activity in T lymphocytes from patients with systemic lupus erythematosus is associated with decreased p65-RelA protein expression. J. Immunol. 1999, 163, 1682–1689. [Google Scholar]
- Kwon, H.-K.; So, J.-S.; Lee, C.-G.; Sahoo, A.; Yi, H.-J.; Park, J.-N.; Lim, S.; Hwang, K.-C.; Jun, C.-D.; Chun, J.-S. Foxp3 induces IL-4 gene silencing by affecting nuclear translocation of NFκB and chromatin structure. Mol. Immunol. 2008, 45, 3205–3212. [Google Scholar] [CrossRef]
- Lee, K.-G.; Xu, S.; Wong, E.-T.; Tergaonkar, V.; Lam, K.-P. Bruton’s tyrosine kinase separately regulates NFκB p65RelA activation and cytokine interleukin (IL)-10/IL-12 production in TLR9-stimulated B cells. J. Biol. Chem. 2008, 283, 11189–11198. [Google Scholar] [CrossRef] [Green Version]








| Surface Antigen | Clone | Source |
|---|---|---|
| CD11c | N418 | eBioscience |
| MHC class II | M5/114.15.2 | eBioscience |
| B220 | RA3-6B2 | eBioscience |
| CD40 | 3/23 | Biolegend |
| CD68 | FA-11 | Biolegend |
| Primer | Primer Sequences | |
|---|---|---|
| Forward | Reverse | |
| β-actin | AAGTGTGACGTTGACATCCGTAAAG | CAGCTCAGTAACAGTCCGCCTAGA |
| Prkcg | CTCCGACGAACTCTATGCCATCAAG | CCAATGCCAGGACACGCTTCTC |
| Pik3r1 | TGTGGCACAGACTTGGTGTT | TTCTTCCCTTGAGATGTCTCCC |
| Akt1 | CCGCCTGATCAAGTTCTCCT | TTCAGATGATCCATGCGGGG |
| Stat3 | TGTCAGATCACATGGGCTAAAT | GGTCGATGATATTGTCTAGCCA |
| Rela | AGACCCAGGAGTGTTCACAGACC | GTCACCAGGCGAGTTATAGCTTCAG |
| Il4 | TACCAGGAGCCATATCCACGGATG | TGTGGTGTTCTTCGTTGCTGTGAG |
| Il12a | GACCTGTTTACCACTGGAACTA | GATCTGCTGATGGTTGTGATTC |
| Cd68 | GAAATGTCACAGTTCACACCAG | GGATCTTGGACTAGTAGCAGTG |
| Ptprc | GTTATCCACGCTGCTGCCTCAC | TTGGCTGCTGAATGTCTGAGTGTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Luo, J.; Du, H.; Liu, G.; Liu, M.; Wang, J.; Han, S.; Che, H. Widely Targeted Lipidomics and Transcriptomics Analysis Revealed Changes of Lipid Metabolism in Spleen Dendritic Cells in Shrimp Allergy. Foods 2022, 11, 1882. https://doi.org/10.3390/foods11131882
Sun S, Luo J, Du H, Liu G, Liu M, Wang J, Han S, Che H. Widely Targeted Lipidomics and Transcriptomics Analysis Revealed Changes of Lipid Metabolism in Spleen Dendritic Cells in Shrimp Allergy. Foods. 2022; 11(13):1882. https://doi.org/10.3390/foods11131882
Chicago/Turabian StyleSun, Shanfeng, Jiangzuo Luo, Hang Du, Guirong Liu, Manman Liu, Junjuan Wang, Shiwen Han, and Huilian Che. 2022. "Widely Targeted Lipidomics and Transcriptomics Analysis Revealed Changes of Lipid Metabolism in Spleen Dendritic Cells in Shrimp Allergy" Foods 11, no. 13: 1882. https://doi.org/10.3390/foods11131882
APA StyleSun, S., Luo, J., Du, H., Liu, G., Liu, M., Wang, J., Han, S., & Che, H. (2022). Widely Targeted Lipidomics and Transcriptomics Analysis Revealed Changes of Lipid Metabolism in Spleen Dendritic Cells in Shrimp Allergy. Foods, 11(13), 1882. https://doi.org/10.3390/foods11131882