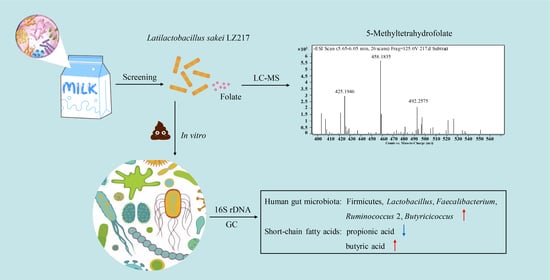
Probiotic Potential of a Folate-Producing Strain Latilactobacillus sakei LZ217 and Its Modulation Effects on Human Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Folate-Producing Lactobacillus Strains
2.2. Analysis of 5-Methyltetrahydrofolate by LC-MS
2.3. Environmental Stress Tolerance Assay
2.4. Simulated Gastrointestinal Tolerance Assay
2.5. The In Vitro Effects of Lactobacillus on Human Gut Microbiota in the Fecal Slurry Cultures
2.5.1. Study Design and Sampling
2.5.2. DNA Extraction and 16S rDNA Gene Sequencing
2.5.3. Detection of Short-Chain Fatty Acids by Gas Chromatography
2.5.4. Statistical Analysis
3. Results and Discussion
3.1. Isolation of Folate-Producing Lactobacillus Strains
3.2. Analysis of the Folate Forms by LC-MS
3.3. Environmental Stress Tolerance Assay
3.4. Simulated Gastrointestinal Tolerance Assay
3.5. Evolution of the Gut Microbiota and the SCFAs Concentrations in the Fecal Slurry Cultures
3.5.1. In Vitro Effect of the L. sakei LZ217 on the Structure of Gut Microbiota
3.5.2. Effect of LZ217 on Short Chain Fatty Acid Content in the Fecal Slurry Cultures
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jacob, R.A. Folate, DNA methylation, and gene expression: Factors of nature and nurture. Am. J. Clin. Nutr. 2000, 72, 903–904. [Google Scholar] [CrossRef] [Green Version]
- Brattström, L.; Wilcken, D.E. Homocysteine and cardiovascular disease: Cause or effect? Am. J. Clin. Nutr. 2000, 72, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Wang, L.; Chen, W.; Menard, K.; Hong, Y.; Tian, Y.; Bonacorsi, S.J.; Humphreys, W.G.; Lee, F.Y.; Gan, J. Tissue distribution and tumor uptake of folate receptor-targeted epothilone folate conjugate, BMS-753493, in CD2F1 mice after systemic administration. Acta Pharm. Sin. B 2016, 6, 460–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Cantley, L.C. Toward a better understanding of folate metabolism in health and disease. J. Exp. Med. 2019, 216, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.G.; Li, Y.L.; Gao, R.F.; Geng, Y.Q.; Chen, X.M.; Liu, X.Q.; Ding, Y.B.; Mu, X.Y.; Wang, Y.X.; He, J.L. Folate deficiency decreases apoptosis of endometrium decidual cells in pregnant mice via the mitochondrial pathway. Nutrients 2015, 7, 1916–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, M.; Amaretti, A.; Raimondi, S. Folate production by probiotic bacteria. Nutrients 2011, 3, 118–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donnelly, J.G. Folic acid. Crit. Rev. Clin. Lab. Sci. 2001, 38, 183–223. [Google Scholar] [CrossRef]
- Ferrazzi, E.; Tiso, G.; Di Martino, D. Folic acid versus 5- methyl tetrahydrofolate supplementation in pregnancy. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 253, 312–319. [Google Scholar] [CrossRef]
- Scaglione, F.; Panzavolta, G. Folate, folic acid and 5-methyltetrahydrofolate are not the same thing. Xenobiotica 2014, 44, 480–488. [Google Scholar] [CrossRef]
- Tamura, T. Determination of food folate. J. Nutr. Biochem. 1998, 9, 285–293. [Google Scholar] [CrossRef]
- Araya-Farias, M.; Gaudreau, A.; Rozoy, E.; Bazinet, L. Rapid HPLC-MS method for the simultaneous determination of tea catechins and folates. J. Agric. Food Chem. 2014, 62, 4241–4250. [Google Scholar] [CrossRef] [PubMed]
- Sybesma, W.; Starrenburg, M.; Tijsseling, L.; Hoefnagel, M.H.; Hugenholtz, J. Effects of cultivation conditions on folate production by lactic acid bacteria. Appl. Environ. Microbiol. 2003, 69, 4542–4548. [Google Scholar] [CrossRef] [Green Version]
- Mo, H.; Kariluoto, S.; Piironen, V.; Zhu, Y.; Sanders, M.G.; Vincken, J.P.; Wolkers-Rooijackers, J.; Nout, M.J. Effect of soybean processing on content and bioaccessibility of folate, vitamin B12 and isoflavones in tofu and tempe. Food Chem. 2013, 141, 2418–2425. [Google Scholar] [CrossRef]
- Jones, M.L.; Nixon, P.F. Tetrahydrofolates are greatly stabilized by binding to bovine milk folate-binding protein. J. Nutr. 2002, 132, 2690–2694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crittenden, R.G.; Martinez, N.R.; Playne, M.J. Synthesis and utilisation of folate by yoghurt starter cultures and probiotic bacteria. Int. J. Food Microbiol. 2003, 80, 217–222. [Google Scholar] [CrossRef]
- Im, S.-H.; Kang, H.-J. Probiotics as an Immune Modulator. J. Nutr. Sci. Vitaminol. 2015, 61, S103–S105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Cai, D.; Yang, M.; Hao, Y.; Zhu, Y.; Chen, Z.; Aziz, T.; Sarwar, A.; Yang, Z. Screening of folate-producing lactic acid bacteria and modulatory effects of folate-biofortified yogurt on gut dysbacteriosis of folate-deficient rats. Food Funct. 2020, 11, 6308–6318. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Factories 2017, 16, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alander, M.; Smet, I.D.; Nollet, L.; Verstraete, W.; Mattila-Sandholm, T.D. The effect of probiotic strains on the microbiota of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME). Int. J. Food Microbiol. 1999, 46, 71–79. [Google Scholar] [CrossRef]
- Li, P.; Zhou, Q.; Gu, Q. Complete genome sequence of Lactobacillus plantarum LZ227, a potential probiotic strain producing B-group vitamins. J. Biotechnol. 2016, 234, 66–70. [Google Scholar] [CrossRef] [Green Version]
- Koontz, J.L.; Phillips, K.M.; Wunderlich, K.M.; Exler, J.; Holden, J.M.; Gebhardt, S.E.; Haytowitz, D.B. Comparison of total folate concentrations in foods determined by microbiological assay at several experienced U.S. commercial laboratories. J. AOAC Int. 2005, 88, 805–813. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Huang, J.; Zhou, R. Progress in engineering acid stress resistance of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2014, 98, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Gu, Q.; Yang, L.; Yu, Y.; Wang, Y. Characterization of extracellular vitamin B12 producing Lactobacillus plantarum strains and assessment of the probiotic potentials. Food Chem. 2017, 234, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Rycroft, C.E.; Jones, M.R.; Gibson, G.R.; Rastall, R.A. Fermentation properties of gentio-oligosaccharides. Lett. Appl. Microbiol. 2010, 32, 156–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, F.; Yin, Y.; Wang, Y.; Deng, B.; Yu, H.D.; Li, L.; Xiang, C.; Wang, S.; Zhu, B.; Wang, X. Higher-level production of volatile fatty acids in vitro by chicken gut microbiotas than by human gut microbiotas as determined by functional analyses. Appl. Environ. Microbiol. 2012, 78, 5763–5772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdés-Varela, L.; Hernández-Barranco, A.M.; Ruas-Madiedo, P.; Gueimonde, M. Effect of Bifidobacterium upon Clostridium difficile Growth and Toxicity When Co-cultured in Different Prebiotic Substrates. Front. Microbiol. 2016, 7, 738. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yan, Y.; Mi, J.; Zhang, H.; Lu, L.; Luo, Q.; Li, X.; Zeng, X.; Cao, Y. Simulated Digestion and Fermentation in Vitro by Human Gut Microbiota of Polysaccharides from Bee Collected Pollen of Chinese Wolfberry. J. Agric. Food Chem. 2018, 66, 898–907. [Google Scholar] [CrossRef]
- Lopes, R.P.; Mota, M.J.; Sousa, S.; Gomes, A.M.; Delgadillo, I.; Saraiva, J.A. Combined effect of pressure and temperature for yogurt production. Food Res. Int. 2019, 122, 222–229. [Google Scholar] [CrossRef]
- Mohd Adnan, A.F.; Tan, I.K. Isolation of lactic acid bacteria from Malaysian foods and assessment of the isolates for industrial potential. Bioresour. Technol. 2007, 98, 1380–1385. [Google Scholar] [CrossRef]
- Masuda, M.; Ide, M.; Utsumi, H.; Niiro, T.; Shimamura, Y.; Murata, M. Production potency of folate, vitamin B12, and thiamine by lactic acid bacteria isolated from Japanese pickles. Biosci. Biotechnol. Biochem. 2012, 76, 2061–2067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todorov, S.D.; Dicks, L.M.T. Evaluation of lactic acid bacteria from kefir, molasses and olive brine as possible probiotics based on physiological properties. Ann. Microbiol. 2008, 58, 661. [Google Scholar] [CrossRef]
- Rupa, P.; Mine, Y. Recent advances in the role of probiotics in human inflammation and gut health. J. Agric. Food Chem. 2012, 60, 8249–8256. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Lin, P.R.; Ng, C.C.; Shyu, Y.T. Probiotic properties of Lactobacillus strains isolated from the feces of breast-fed infants and Taiwanese pickled cabbage. Anaerobe 2010, 16, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 1998, 84, 759–768. [Google Scholar] [CrossRef]
- Hang, S.T.; Zeng, L.z.; Han, J.r.; Zhang, Z.q.; Zhou, Q.; Meng, X.; Gu, Q.; Li, P. Lactobacillus plantarum ZJ316 improves the quality of Stachys sieboldii Miq. pickle by inhibiting harmful bacteria growth and degrading nitrite, promoting the gut microbiota health in vitro. Food Funct. 2022; accepted manuscript. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Silk, D.B.; Davis, A.; Vulevic, J.; Tzortzis, G.; Gibson, G.R. Clinical trial: The effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment. Pharmacol. Ther. 2009, 29, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Engevik, M.A.; Morra, C.N.; Röth, D.; Engevik, K.; Spinler, J.K.; Devaraj, S.; Crawford, S.E.; Estes, M.K.; Kalkum, M.; Versalovic, J. Microbial Metabolic Capacity for Intestinal Folate Production and Modulation of Host Folate Receptors. Front. Microbiol. 2019, 10, 2305. [Google Scholar] [CrossRef] [PubMed]
- Eeckhaut, V.; Van Immerseel, F.; Teirlynck, E.; Pasmans, F.; Fievez, V.; Snauwaert, C.; Haesebrouck, F.; Ducatelle, R.; Louis, P.; Vandamme, P. Butyricicoccus pullicaecorum gen. nov., sp. nov., an anaerobic, butyrate-producing bacterium isolated from the caecal content of a broiler chicken. Int. J. Syst. Evol. Microbiol. 2008, 58, 2799–2802. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Li, S.; Luo, J.; Wu, X.; Liu, L. Effects of dietary fibers and their mixtures on short chain fatty acids and microbiota in mice guts. Food Funct. 2013, 4, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, E.; Grootaert, C.; Verstraete, W.; Van de Wiele, T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 2011, 69, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.V.; Frassetto, A.; Kowalik, E.J., Jr.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G.; et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef] [PubMed]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854.e1. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Si, Y.; Xu, J.; Lin, Y.; Wang, J.Z.; Cao, M.; Sun, S.; Ding, Q.; Zhu, L.; Wei, J.F. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J. Cell. Mol. Med. 2020, 24, 3521–3533. [Google Scholar] [CrossRef] [Green Version]
- Spalinger, M.R.; Schmidt, T.S.; Schwarzfischer, M.; Hering, L.; Atrott, K.; Lang, S.; Gottier, C.; Geirnaert, A.; Lacroix, C.; Dai, X.; et al. Protein tyrosine phosphatase non-receptor type 22 modulates colitis in a microbiota-dependent manner. J. Clin. Investig. 2019, 129, 2527–2541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Liu, Y.; Duan, X.; Xiao, C.; Lan, Y.; Luo, L.; Wu, C.; Yang, Z.; Mai, X.; Lu, S.; et al. Alteration of the gut microbiota by vinegar is associated with amelioration of hyperoxaluria-induced kidney injury. Food Funct. 2020, 11, 2639–2653. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Li, Y.; Han, H.; Chen, S.; Gao, J.; Liu, G.; Wu, X.; Deng, J.; Yu, Q.; Huang, X.; et al. Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice. J. Pineal Res. 2018, 65, e12524. [Google Scholar] [CrossRef]
- Lordan, C.; Thapa, D.; Ross, R.P.; Cotter, P.D. Potential for enriching next-generation health-promoting gut bacteria through prebiotics and other dietary components. Gut Microbes 2020, 11, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.B.; Xiao, L.; Xing, S.C.; Chen, J.Y.; Yang, Y.W.; Zhou, Y.; Chen, W.; Liang, J.B.; Mi, J.D.; Wang, Y. The microbiota structure in the cecum of laying hens contributes to dissimilar H2S production. BMC Genom. 2019, 20, 770. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Chen, Q.; Sun, Y.; Zeng, L.; Wu, H.; Gu, Q.; Li, P. Probiotic Potential of a Folate-Producing Strain Latilactobacillus sakei LZ217 and Its Modulation Effects on Human Gut Microbiota. Foods 2022, 11, 234. https://doi.org/10.3390/foods11020234
Liu M, Chen Q, Sun Y, Zeng L, Wu H, Gu Q, Li P. Probiotic Potential of a Folate-Producing Strain Latilactobacillus sakei LZ217 and Its Modulation Effects on Human Gut Microbiota. Foods. 2022; 11(2):234. https://doi.org/10.3390/foods11020234
Chicago/Turabian StyleLiu, Manman, Qingqing Chen, Yalian Sun, Lingzhou Zeng, Hongchen Wu, Qing Gu, and Ping Li. 2022. "Probiotic Potential of a Folate-Producing Strain Latilactobacillus sakei LZ217 and Its Modulation Effects on Human Gut Microbiota" Foods 11, no. 2: 234. https://doi.org/10.3390/foods11020234
APA StyleLiu, M., Chen, Q., Sun, Y., Zeng, L., Wu, H., Gu, Q., & Li, P. (2022). Probiotic Potential of a Folate-Producing Strain Latilactobacillus sakei LZ217 and Its Modulation Effects on Human Gut Microbiota. Foods, 11(2), 234. https://doi.org/10.3390/foods11020234