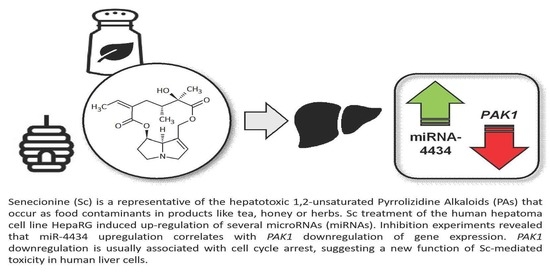
Identification of microRNAs Implicated in Modulating Senecionine-Induced Liver Toxicity in HepaRG Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cell Culture
2.3. Liver-Specific miRNA Array
2.4. RNA Isolation and qPCR Analysis
2.5. Ingenuity Pathway Analysis
2.6. AntagomiR Experiments
2.7. Cell Viability Assay
2.8. Statistical Analysis
3. Results
3.1. Sc Induces miRNA Expression in HepaRG Cells
3.2. Prediction of Biological Consequences of PA-Mediated miRNA Level Alterations
3.3. MiR-4434 Negatively Regulates PAK1 Gene Expression in HepaRG Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALARA | As Low As Reasonably Achievable |
| ATP6V1H/0E2 | V-type proton ATPase subunit H/E 2 |
| DEG | Differentially Expressed Gene |
| DMSO | DiMethyl SulfOxide |
| FBS | Fetal Bovine Serum |
| FC | Fold Change |
| FDR | False Discovery Rate |
| GAS2 | Growth Arrest-Specific protein 2 |
| GUSB | β-glucuronidase |
| HSOS | Hepatic Sinusoidal Obstruction Syndrome |
| IPA | Ingenuity Pathway Analysis |
| Lc | Lasiocarpine |
| lncRNA | long non-coding RNA |
| LSINCT5 | Long Stress-Induced Non-Coding Transcripts 5 |
| LEPROT | LEPtin Receptor Overlapping Transcript |
| Mc | Monocrotaline |
| miRNA | microRNA |
| NRU | Neutral Red Uptake |
| PA | 1,2-unsaturated PyrrolizidineAalkaloid |
| PAK1 | P21-Activated Kinase-1 |
| PATZ1 | POZ/BTB and AT hook containing Zinc finger 1 |
| PRKAR1A/2A | cAMP-dependent PRotein Kinase type I (II)-Alpha Regulatory subunit |
| PXR | Pregnane X Receptor |
| qPCR | quantitative real-time Polymerase Chain Reaction |
| RT | Room Temperature |
| Sc | Senecionine |
| ST6GAL1 | ST6 beta-GALactoside alpha-2,6-sialyltransferase 1 |
| TDI | Tolerable Daily Intake |
| UTR | 3′-UnTranslated Region |
References
- Mattocks, A. Chemistry and Toxicology of Pyrrolizidine Alkaloids; Academic Press: London, UK, 1986. [Google Scholar]
- Mulder, P.P.J.; López, P.; Castellari, M.; Bodi, D.; Ronczka, S.; Preiss-Weigert, A.; These, A. Occurrence of pyrrolizidine alkaloids in animal- and plant-derived food: Results of a survey across Europe. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2018, 35, 118–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodi, D.; Ronczka, S.; Gottschalk, C.; Behr, N.; Skibba, A.; Wagner, M.; Lahrssen-Wiederholt, M.; Preiss-Weigert, A.; These, A. Determination of pyrrolizidine alkaloids in tea, herbal drugs and honey. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2014, 31, 1886–1895. [Google Scholar] [CrossRef] [PubMed]
- Edgar, J.A.; Lin, H.J.; Kumana, C.R.; Ng, M.M. Pyrrolizidine alkaloid composition of three Chinese medicinal herbs, Eupatorium cannabinum, E. japonicum and Crotalaria assamica. Am. J. Chin. Med. 1992, 20, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Kaltner, F.; Rychlik, M.; Gareis, M.; Gottschalk, C. Occurrence and Risk Assessment of Pyrrolizidine Alkaloids in Spices and Culinary Herbs from Various Geographical Origins. Toxins 2020, 12, 155. [Google Scholar] [CrossRef] [Green Version]
- BfR. Pyrrolizidine Alkaloids in Herbal Teas and Teas; BfR Opinion No. 018/2013; German Federal Institute for Risk Assessment: Berlin, Germany, 2013; pp. 1–31.
- Ruan, J.; Yang, M.; Fu, P.; Ye, Y.; Lin, G. Metabolic activation of pyrrolizidine alkaloids: Insights into the structural and enzymatic basis. Chem. Res. Toxicol. 2014, 27, 1030–1039. [Google Scholar] [CrossRef]
- Bach, N.; Thung, S.N.; Schaffner, F. Comfrey herb tea-induced hepatic veno-occlusive disease. Am. J. Med. 1989, 87, 97–99. [Google Scholar] [CrossRef]
- Ridker, P.M.; McDermott, W.V. Comfrey herb tea and hepatic veno-occlusive disease. Lancet 1989, 1, 657–658. [Google Scholar] [CrossRef]
- Ridker, P.M.; Ohkuma, S.; McDermott, W.V.; Trey, C.; Huxtable, R.J. Hepatic venocclusive disease associated with the consumption of pyrrolizidine-containing dietary supplements. Gastroenterology 1985, 88, 1050–1054. [Google Scholar] [CrossRef]
- Kumana, C.R.; Ng, M.; Lin, H.J.; Ko, W.; Wu, P.C.; Todd, D. Herbal tea induced hepatic veno-occlusive disease: Quantification of toxic alkaloid exposure in adults. Gut 1985, 26, 101–104. [Google Scholar] [CrossRef] [Green Version]
- Roitman, J.N. Comfrey and liver damage. Lancet 1981, 1, 944. [Google Scholar] [CrossRef]
- Edgar, J.A.; Molyneux, R.J.; Colegate, S.M. Pyrrolizidine Alkaloids: Potential Role in the Etiology of Cancers, Pulmonary Hypertension, Congenital Anomalies, and Liver Disease. Chem. Res. Toxicol. 2015, 28, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Edgar, J.A.; Roeder, E.; Molyneux, R.J. Honey from plants containing pyrrolizidine alkaloids: A potential threat to health. J. Agric. Food Chem. 2002, 50, 2719–2730. [Google Scholar] [CrossRef]
- Svoboda, D.; Reddy, J.; Bunyaratvej, S. Hepatic megalocytosis in chronic lasiocarpine poisoning. Some functional studies. Am. J. Pathol 1971, 65, 399–409. [Google Scholar]
- Jago, M.V. The development of the hepatic megalocytosis of chronic pyrrolizidine alkaloid poisoning. Am. J. Pathol. 1969, 56, 405–421. [Google Scholar]
- Molteni, A.; Ward, W.F.; Ts’Ao, C.-H.; Port, C.D.; Solliday, N.H. Monocrotaline-lnduced Pulmonary Endothelial Dysfunction in Rats. Proc. Soc. Exp. Biol. Med. 1984, 176, 88–94. [Google Scholar] [CrossRef]
- Stegelmeier, B.L.; Edgar, J.A.; Colegate, S.M.; Gardner, D.R.; Schoch, T.K.; Coulombe, R.A.; Molyneux, R.J. Pyrrolizidine alkaloid plants, metabolism and toxicity. J. Nat. Toxins 1999, 8, 95–116. [Google Scholar] [PubMed]
- Waizenegger, J.; Braeuning, A.; Templin, M.; Lampen, A.; Hessel-Pras, S. Structure-dependent induction of apoptosis by hepatotoxic pyrrolizidine alkaloids in the human hepatoma cell line HepaRG: Single versus repeated exposure. Food Chem. Toxicol. 2018, 114, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Ebmeyer, J.; Braeuning, A.; Glatt, H.; These, A.; Hessel-Pras, S.; Lampen, A. Human CYP3A4-mediated toxification of the pyrrolizidine alkaloid lasiocarpine. Food Chem. Toxicol. 2019, 130, 79–88. [Google Scholar] [CrossRef]
- Glück, J.; Waizenegger, J.; Braeuning, A.; Hessel-Pras, S. Pyrrolizidine Alkaloids Induce Cell Death in Human HepaRG Cells in a Structure-Dependent Manner. Int. J. Mol. Sci. 2021, 22, 202. [Google Scholar] [CrossRef] [PubMed]
- Allemang, A.; Mahony, C.; Lester, C.; Pfuhler, S. Relative potency of fifteen pyrrolizidine alkaloids to induce DNA damage as measured by micronucleus induction in HepaRG human liver cells. Food Chem. Toxicol. 2018, 121, 72–81. [Google Scholar] [CrossRef]
- Glück, J.; Henricsson, M.; Braeuning, A.; Hessel-Pras, S. The Food Contaminants Pyrrolizidine Alkaloids Disturb Bile Acid Homeostasis Structure-Dependently in the Human Hepatoma Cell Line HepaRG. Foods 2021, 10, 1114. [Google Scholar] [CrossRef] [PubMed]
- Waizenegger, J.; Glück, J.; Henricsson, M.; Luckert, C.; Braeuning, A.; Hessel-Pras, S. Pyrrolizidine Alkaloids Disturb Bile Acid Homeostasis in the Human Hepatoma Cell Line HepaRG. Foods 2021, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Hessel-Pras, S.; Braeuning, A.; Guenther, G.; Adawy, A.; Enge, A.-M.; Ebmeyer, J.; Henderson, C.J.; Hengstler, J.G.; Lampen, A.; Reif, R. The pyrrolizidine alkaloid senecionine induces CYP-dependent destruction of sinusoidal endothelial cells and cholestasis in mice. Arch. Toxicol. 2020, 94, 219–229. [Google Scholar] [CrossRef]
- Luckert, C.; Braeuning, A.; Lampen, A.; Hessel-Pras, S. PXR: Structure-specific activation by hepatotoxic pyrrolizidine alkaloids. Chem. Biol. Interact. 2018, 288, 38–48. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Oda, S.; Tsuneyama, K.; Yokoi, T. Comprehensive analysis of serum microRNAs in hepatic sinusoidal obstruction syndrome (SOS) in rats: Implication as early phase biomarkers for SOS. Arch. Toxicol. 2018, 92, 2947–2962. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Munoz-Garrido, P.; García-Fernández de Barrena, M.; Hijona, E.; Carracedo, M.; Marín, J.J.G.; Bujanda, L.; Banales, J.M. MicroRNAs in biliary diseases. World J. Gastroenterol. 2012, 18, 6189–6196. [Google Scholar] [CrossRef]
- Garzon, R.; Marcucci, G. Potential of microRNAs for cancer diagnostics, prognostication and therapy. Curr. Opin. Oncol. 2012, 24, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Szabo, G.; Bala, S. MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 542–552. [Google Scholar] [CrossRef] [Green Version]
- Marin, J.J.; Bujanda, L.; Banales, J.M. MicroRNAs and cholestatic liver diseases. Curr. Opin. Gastroenterol. 2014, 30, 303–309. [Google Scholar] [CrossRef]
- Song, K.H.; Li, T.; Owsley, E.; Chiang, J.Y. A putative role of micro RNA in regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes. J. Lipid Res. 2010, 51, 2223–2233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Li, Z.; Yan, J.; Yang, X.; Salminen, W. MicroRNA expression profiles distinguish the carcinogenic effects of riddelliine in rat liver. Mutagenesis 2012, 27, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Chen, M.; Zhang, J.; Sheng, Y.; Ji, L. Integrative analysis of hepatic microRNA and mRNA to identify potential biological pathways associated with monocrotaline-induced liver injury in mice. Toxicol. Appl. Pharmacol. 2017, 333, 35–42. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, W.; Yang, Y.; Chen, Y.; Zhuge, Y.; Xiong, A.; Yang, L.; Wang, Z. Blood microRNA Signatures Serve as Potential Diagnostic Biomarkers for Hepatic Sinusoidal Obstruction Syndrome Caused by Gynura japonica Containing Pyrrolizidine Alkaloids. Front. Pharmacol. 2021, 12, 627126. [Google Scholar] [CrossRef]
- Oda, S.; Takeuchi, M.; Akai, S.; Shirai, Y.; Tsuneyama, K.; Yokoi, T. miRNA in Rat Liver Sinusoidal Endothelial Cells and Hepatocytes and Application to Circulating Biomarkers that Discern Pathogenesis of Liver Injuries. Am. J. Pathol. 2018, 188, 916–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gripon, P.; Rumin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 2002, 99, 15655–15660. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. miRecords: An integrated resource for microRNA–target interactions. Nucleic Acids Res. 2009, 37, D105–D110. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Luckert, C.; Hessel, S.; Lenze, D.; Lampen, A. Disturbance of gene expression in primary human hepatocytes by hepatotoxic pyrrolizidine alkaloids: A whole genome transcriptome analysis. Toxicol. Vitr. 2015, 29, 1669–1682. [Google Scholar] [CrossRef]
- Enge, A.M.; Kaltner, F.; Gottschalk, C.; Kin, A.; Kirstgen, M.; Geyer, J.; These, A.; Hammer, H.; Pötz, O.; Braeuning, A.; et al. Organic Cation Transporter I and Na+ /taurocholate Co-Transporting Polypeptide are Involved in Retrorsine- and Senecionine-Induced Hepatotoxicity in HepaRG cells. Mol. Nutr. Food Res. 2021, e2100800. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Sheedy, F.J.; McCoy, C.E. MicroRNAs: The fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 2011, 11, 163–175. [Google Scholar] [CrossRef]
- Bala, S.; Szabo, G. MicroRNA Signature in Alcoholic Liver Disease. Int. J. Hepatol. 2012, 2012, 498232. [Google Scholar] [CrossRef] [Green Version]
- Thai, T.H.; Calado, D.P.; Casola, S.; Ansel, K.M.; Xiao, C.; Xue, Y.; Murphy, A.; Frendewey, D.; Valenzuela, D.; Kutok, J.L.; et al. Regulation of the germinal center response by microRNA-155. Science 2007, 316, 604–608. [Google Scholar] [CrossRef]
- An, F.; Gong, B.; Wang, H.; Yu, D.; Zhao, G.; Lin, L.; Tang, W.; Yu, H.; Bao, S.; Xie, Q. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis 2012, 17, 702–716. [Google Scholar] [CrossRef]
- Yu, D.S.; An, F.M.; Gong, B.D.; Xiang, X.G.; Lin, L.Y.; Wang, H.; Xie, Q. The regulatory role of microRNA-1187 in TNF-α-mediated hepatocyte apoptosis in acute liver failure. Int. J. Mol. Med. 2012, 29, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.D.; Narain, N.; Händel, E.M.; Iken, M.; Singhal, N.; Cathomen, T.; Manns, M.P.; Schöler, H.R.; Ott, M.; Cantz, T. MicroRNA-221 regulates FAS-induced fulminant liver failure. Hepatology 2011, 53, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Loya, K.; Rani, B.; Möbus, S.; Balakrishnan, A.; Lamle, J.; Cathomen, T.; Vogel, A.; Manns, M.P.; Ott, M.; et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology 2013, 57, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Noetel, A.; Kwiecinski, M.; Elfimova, N.; Huang, J.; Odenthal, M. microRNA are Central Players in Anti- and Profibrotic Gene Regulation during Liver Fibrosis. Front. Physiol. 2012, 3, 49. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Huang, C.; Zhang, S.P.; Sun, X.; Long, X.R.; Li, J. The potential of microRNAs in liver fibrosis. Cell Signal 2012, 24, 2268–2272. [Google Scholar] [CrossRef]
- Hu, W.; Coller, J. What comes first: Translational repression or mRNA degradation? The deepening mystery of microRNA function. Cell Res. 2012, 22, 1322–1324. [Google Scholar] [CrossRef] [Green Version]
- Djuranovic, S.; Nahvi, A.; Green, R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 2012, 336, 237–240. [Google Scholar] [CrossRef] [Green Version]
- Bazzini, A.A.; Lee, M.T.; Giraldez, A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012, 336, 233–237. [Google Scholar] [CrossRef] [Green Version]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef] [Green Version]
- Gholipour, N.; Ohradanova-Repic, A.; Ahangari, G. A novel report of MiR-4301 induces cell apoptosis by negatively regulating DRD2 expression in human breast cancer cells. J. Cell Biochem. 2018, 119, 6408–6417. [Google Scholar] [CrossRef] [PubMed]
- Avval, A.J.; Majd, A.; Gholipour, N.; Noghabi, K.A.; Ohradanova-Repic, A.; Ahangari, G. An Inventive Report of Inducing Apoptosis in Non-Small Cell Lung Cancer (NSCLC) Cell Lines by Transfection of MiR-4301. Anticancer Agents Med. Chem. 2019, 19, 1609–1617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Li, H.; Wang, G.X.; Wang, J.; Xiang, Y.; Huang, Y.; Shen, C.; Dai, Z.T.; Li, J.P.; Zhang, T.C.; et al. MKL1/miR-5100/CAAP1 loop regulates autophagy and apoptosis in gastric cancer cells. Neoplasia 2020, 22, 220–230. [Google Scholar] [CrossRef]
- Yang, L.; Lin, Z.; Wang, Y.; Gao, S.; Li, Q.; Li, C.; Xu, W.; Chen, J.; Liu, T.; Song, Z.; et al. MiR-5100 increases the cisplatin resistance of the lung cancer stem cells by inhibiting the Rab6. Mol. Carcinog. 2018, 57, 419–428. [Google Scholar] [CrossRef]
- Lin, H.; Zhang, R.; Wu, W.; Lei, L. miR-4454 Promotes Hepatic Carcinoma Progression by Targeting Vps4A and Rab27A. Oxid. Med. Cell Longev. 2021, 2021, 9230435. [Google Scholar] [CrossRef]
- Ye, D.; Zhang, T.; Lou, G.; Liu, Y. Role of miR-223 in the pathophysiology of liver diseases. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Li, J.; Bie, B.; Sun, J.; Mu, Y.; Shi, M.; Zhang, S.; Kong, G.; Li, Z.; Guo, Y. MiR-3663-3p participates in the anti-hepatocellular carcinoma proliferation activity of baicalein by targeting SH3GL1 and negatively regulating EGFR/ERK/NF-κB signaling. Toxicol. Appl. Pharmacol. 2021, 420, 115522. [Google Scholar] [CrossRef] [PubMed]
- National Toxicology Program. Bioassay of lasiocarpine for possible carcinogenicity. Natl. Cancer Inst. Carcinog. Tech. Rep. Ser. 1978, 39, 1–66. [Google Scholar]
- National Toxicology Program. Toxicology and carcinogenesis studies of riddelliine (CAS No. 23246-96-0) in F344/N rats and B6C3F1 mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 2003, 508, 1–280. [Google Scholar]
- Chen, T.; Mei, N.; Fu, P.P. Genotoxicity of pyrrolizidine alkaloids. J. Appl. Toxicol. 2010, 30, 183–196. [Google Scholar] [CrossRef]
- Jin, X.H.; Lu, S.; Wang, A.F. Expression and clinical significance of miR-4516 and miR-21-5p in serum of patients with colorectal cancer. BMC Cancer 2020, 20, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, T.; Bell, E.H.; McElroy, J.; Becker, A.P.; Gulati, P.M.; Geurts, M.; Mladkova, N.; Gray, A.; Liu, K.; Yang, L.; et al. miR-4516 predicts poor prognosis and functions as a novel oncogene via targeting PTPN14 in human glioblastoma. Oncogene 2019, 38, 2923–2936. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xu, M.; Zhao, J.; Shen, J.; Li, J.; Liu, Y.; Cao, G.; Ma, J.; He, W.; Chen, X.; et al. MicroRNA-4516 suppresses pancreatic cancer development via negatively regulating orthodenticle homeobox 1. Int. J. Biol. Sci. 2020, 16, 2159–2169. [Google Scholar] [CrossRef]
- Li, O.; Li, Z.; Tang, Q.; Li, Y.; Yuan, S.; Shen, Y.; Zhang, Z.; Li, N.; Chu, K.; Lei, G. Long Stress Induced Non-Coding Transcripts 5 (LSINCT5) Promotes Hepatocellular Carcinoma Progression Through Interaction with High-Mobility Group AT-hook 2 and MiR-4516. Med. Sci. Monit. 2018, 24, 8510–8523. [Google Scholar] [CrossRef]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [Green Version]
- Van Keuren-Jensen, K.R.; Malenica, I.; Courtright, A.L.; Ghaffari, L.T.; Starr, A.P.; Metpally, R.P.; Beecroft, T.A.; Carlson, E.W.; Kiefer, J.A.; Pockros, P.J.; et al. microRNA changes in liver tissue associated with fibrosis progression in patients with hepatitis C. Liver Int. 2016, 36, 334–343. [Google Scholar] [CrossRef] [Green Version]
- Rane, C.K.; Minden, A. P21 activated kinase signaling in cancer. Semin. Cancer Biol. 2019, 54, 40–49. [Google Scholar] [CrossRef]
- Radu, M.; Semenova, G.; Kosoff, R.; Chernoff, J. PAK signalling during the development and progression of cancer. Nat. Rev. Cancer 2014, 14, 13–25. [Google Scholar] [CrossRef]
- Advani, S.J.; Camargo, M.F.; Seguin, L.; Mielgo, A.; Anand, S.; Hicks, A.M.; Aguilera, J.; Franovic, A.; Weis, S.M.; Cheresh, D.A. Kinase-independent role for CRAF-driving tumour radioresistance via CHK2. Nat. Commun. 2015, 6, 8154. [Google Scholar] [CrossRef] [Green Version]
- Arias-Romero, L.E.; Chernoff, J. A tale of two Paks. Biol. Cell 2008, 100, 97–108. [Google Scholar] [CrossRef]
- Tu, J.; Zhao, Z.; Xu, M.; Chen, M.; Weng, Q.; Ji, J. LINC00460 promotes hepatocellular carcinoma development through sponging miR-485-5p to up-regulate PAK1. Biomed. Pharmacother. 2019, 118, 109213. [Google Scholar] [CrossRef]
- Abdelfatah, S.; Naß, J.; Knorz, C.; Klauck, S.M.; Küpper, J.H.; Efferth, T. Pyrrolizidine alkaloids cause cell cycle and DNA damage repair defects as analyzed by transcriptomics in cytochrome P450 3A4-overexpressing HepG2 clone 9 cells. Cell Biol. Toxicol. 2021, 1–21. [Google Scholar] [CrossRef]





| Gene | Ensembl ID | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|---|
| ACVR2A | ENSG00000121989 | TGTTTTGGGCACAGGTTATTGT | CAAGGTGGGGTTTTATGGGGA |
| AOX1 | ENSG00000138356 | TCCCTGCCATCTGTGACATG | CCGACTCTCCCAGACCCTTA |
| ATP6V1H | ENSG00000047249 | CCAAAACTGTGGCCATGCTA | TGCTGGTGAGAGGCTTCAGA |
| ATP7A | ENSG00000165240 | CAAAACCAGGCGTCTCAACC | TCGTCCACTGCTGTTTTCGG |
| BOK | ENSG00000176720 | CACACACAGCCTTCCCTTGA | GCCTGTATCTCCTGAGTGCC |
| CASP2 | ENSG00000106144 | GAATTCCACCGGTGCAAGGA | TGGATGATGGGGAGGTGACA |
| FBXW7 | ENSG00000109670 | TATACTCCCTGCCCTTCCCC | CCAACATCCTGCACCACTGA |
| FOXO1 | ENSG00000150907 | CAGGGGTGGCCATGTAAGTC | GGAACAAGAACGTGGAATCTGC |
| GAS2 | ENSG00000148935 | GTGCCGAGATTTAGGGGTGG | GCAATCCGGCCAAGCTCTAG |
| GUSB | ENSG00000169919 | CACCAGGGACCATCCAATAC | ATGTAGGTGGTGGGTGTCGT |
| IFIH1 | ENSG00000115267 | CAAAAGAAGTGTGCCGACTATCA | TGCACCATCATTGTTCCCCA |
| LEPROT | ENSG00000213625 | ATTGAGTTCCCAGGCCAAGC | AGACCCAAAACTCAGGCAGG |
| PAK1 | ENSG00000149269 | GCCTGACATGATACCCTGCC | AGCAAACATCCCCAACACCC |
| PATZ1 | ENSG00000100105 | AGTCTGGTCAGGGAAGTAGGG | ACACAATGTCCCTACCTGCC |
| PRKAR1A | ENSG00000108946 | ACTTTGCTGGAGTGGTGGTG | TGGTCTTGAACTCCTGGGCT |
| RACGAP1 | ENSG00000161800 | CTGTCCCCTTCCCTGCATTC | TGCACAACAATGGAGGGGAT |
| REL | ENSG00000162924 | CTTCATGCCCCTTCCCAGTC | ACGTTGACAACCCAGCTGTT |
| SFXN1 | ENSG00000164466 | AACAGGGTCATGCTTGGATCA | GGCTTCTTGAGGTCTCTGGC |
| SLC25A29 | ENSG00000197119 | GCCCACACTGTAGAGTCACG | AGAAAGGGGCTGGAGTGTCT |
| SLC4A4 | ENSG00000080493 | CAAACATTGCAACTCAGGGCT | TTCACATTGTAGGACTGGGACA |
| STX17 | ENSG00000136874 | TGTTAGCAAGGGTCAGCACG | TCAAGCACCACTCAGCAATGT |
| ST6GAL1 | ENSG00000073849 | AGCCCACTTTCCCTCTCCAT | TGCCTCTCTCACTGAACCGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enge, A.-M.; Sprenger, H.; Braeuning, A.; Hessel-Pras, S. Identification of microRNAs Implicated in Modulating Senecionine-Induced Liver Toxicity in HepaRG Cells. Foods 2022, 11, 532. https://doi.org/10.3390/foods11040532
Enge A-M, Sprenger H, Braeuning A, Hessel-Pras S. Identification of microRNAs Implicated in Modulating Senecionine-Induced Liver Toxicity in HepaRG Cells. Foods. 2022; 11(4):532. https://doi.org/10.3390/foods11040532
Chicago/Turabian StyleEnge, Anne-Margarethe, Heike Sprenger, Albert Braeuning, and Stefanie Hessel-Pras. 2022. "Identification of microRNAs Implicated in Modulating Senecionine-Induced Liver Toxicity in HepaRG Cells" Foods 11, no. 4: 532. https://doi.org/10.3390/foods11040532
APA StyleEnge, A. -M., Sprenger, H., Braeuning, A., & Hessel-Pras, S. (2022). Identification of microRNAs Implicated in Modulating Senecionine-Induced Liver Toxicity in HepaRG Cells. Foods, 11(4), 532. https://doi.org/10.3390/foods11040532