Crosslinked Recombinant-Ara h 1 Catalyzed by Microbial Transglutaminase: Preparation, Structural Characterization and Allergic Assessment
Abstract
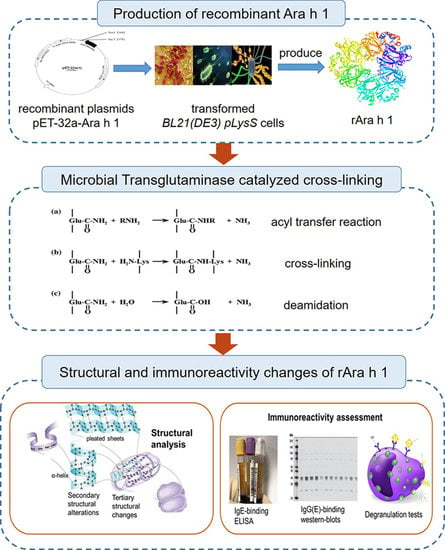
:1. Introduction
2. Materials and Methods
2.1. Material and Reagents
2.2. Human Sera
2.3. Preparation of Cross-Linked rAra h 1
2.4. Determination of Structural Alterations
2.4.1. Polyacrylamide Gel Electrophoresis (PAGE)
2.4.2. Intrinsic Fluorescence Spectroscopy
2.4.3. Dynamic Light Scattering
2.4.4. Determination of the Secondary Bonds
2.4.5. Surface Hydrophobicity (H0) Measurement
2.4.6. Analysis of Protein Secondary Structures
2.4.7. Spatial Structure Analysis
2.5. Immunoreactivity Assessment
2.5.1. Enzyme Linked Immunosorbent Assay (ELISA)
2.5.2. Immunoblotting
2.5.3. Degranulation Test
2.5.4. Statistical Analysis
3. Results and Discussion
3.1. Preparation of Cross-Linked rAra h 1
3.1.1. Influence of Protein Concentration on the rAra h 1 Aggregation
3.1.2. Effect of Reaction Conditions (Temperature, pH, and Time) on the rAra h 1 Aggregation
3.1.3. Effect of Reduction Extent on the rAra h 1 Cross-Linking
3.2. Structural Changes in Polymerized rAra h 1
3.2.1. Intrinsic Fluorescence and Molecular Size
3.2.2. Aggregation Mode, Surface Hydrophobicity and Chemical Bonds
3.2.3. CD Spectra
3.3. Effect of Cross-Linking on the Immunoreactivity
3.3.1. Immunoblots and ELISA Analysis
3.3.2. β-hexosaminidase Release
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Ni, S.; Wang, C.; Li, X.; Fu, L. Cross-linking of shrimp tropomyosin catalyzed by transglutaminase and tyrosinase produces hypoallergens for potential immunotherapy. Food Funct. 2019, 10, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, J.S.; Yang, X.J.; Lin, D.H.; Gao, Y.F.; Su, Y.J.; Yang, S.; Zhang, Y.J.; Zheng, J.J. Peanut Allergy, Allergen Composition, and Methods of Reducing Allergenicity: A Review. Int. J. Food Sci. 2013, 909140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.M.; Sicherer, S.H.; Burks, A.W.; Leung, D.Y.M.; Lindblad, R.W.; Dawson, P.; Henning, A.K.; Berin, M.C.; Chiang, D.; Vickery, B.P. Epicutaneous immunotherapy for the treatment of peanut allergy in children and young adults. J. Allergy Clin. Immunol. 2017, 139, 1242–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burks, W.; Sampson, H.A.; Bannon, G.A. Peanut allergens. Allergy 1998, 53, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Blanc, F.; Vissers, Y.M.; Adel-Patient, K.; Rigby, N.M.; Mackie, A.R.; Gunning, A.P.; Wellner, N.K.; Skov, P.S.; Przybylski-Nicaise, L.; Ballmer-Weber, B.; et al. Boiling peanut Ara h 1 results in the formation of aggregates with reduced allergenicity. Mol. Nutr. Food Res. 2011, 55, 1887–1894. [Google Scholar] [CrossRef]
- Ramesh, M.; Yuenyongviwat, A.; Konstantinou, G.N.; Lieberman, J.; Pascal, M.; Masilamani, M.; Sampson, H.A. Peanut T-cell epitope discovery: Ara h 1. J. Allergy Clin. Immunol. 2016, 137, 1764–1771. [Google Scholar] [CrossRef] [Green Version]
- Maleki, S.J.; Kopper, R.A.; Shin, D.S.; Park, C.W.; Compadre, C.M.; Sampson, H.; Burks, A.W.; Bannon, G.A. Structure of the major peanut allergen Ara h 1 may protect IgE-binding epitopes from degradation. J. Immunol. 2000, 164, 5844–5849. [Google Scholar] [CrossRef] [Green Version]
- Maleki, S. The molecular effects of processing on the peanut allergens. Clin. Transl. Allergy 2014, 4, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Yu, J.; Ahmedna, M.; Goktepe, I. Reduction of major peanut allergens Ara h 1 and Ara h 2, in roasted peanuts by ultrasound assisted enzymatic treatment. Food Chem. 2013, 141, 762–768. [Google Scholar] [CrossRef]
- López, E.; Cuadrado, C.; Burban, C. Effects of autoclaving and high pressure on allergenicity of hazelnut proteins. J. Clin. Bioinform. 2012, 2, 12. [Google Scholar] [CrossRef] [Green Version]
- Cuadrado, C.; Cheng, H.; Sanchiz, A.; Ballesteros, I.; Easson, M.; Grimm, C.C.; Dieguez, M.C.; Linacero, R.; Burbano, C.; Maleki, S.J. Influence of enzymatic hydrolysis on the allergenic reactivity of processed cashew and pistachio. Food Chem. 2018, 241, 372–379. [Google Scholar] [CrossRef]
- Rao, H.; Chen, C.; Tian, Y.; Li, Y.; Gao, Y.; Tao, S.; Xue, W. Germination results in reduced allergenicity of peanut by degradation of allergens and resveratrol enrichment. Innov. Food Sci. Emerg. Technol. 2018, 50, 188–195. [Google Scholar] [CrossRef]
- Fu, W.; Xue, W.; Liu, C.; Tian, Y.; Zhu, Z. Screening of Lactic Acid Bacteria and Yeasts from Sourdough as Starter Cultures for Reduced Allergenicity Wheat Products. Foods 2020, 9, 751. [Google Scholar] [CrossRef]
- Vanga, S.K.; Singh, A.; Raghavan, V. Effect of thermal and electric field treatment on the conformation of Ara h 6 peanut protein allergen. Innov. Food Sci. Emerg. Technol. 2015, 30, 79–88. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, C.; Zhang, K.; Tao, S.; Xue, W. Glycosylation between recombinant peanut protein Ara h 1 and glucosamine could decrease the allergenicity due to the protein aggregation. LWT Food Sci. Technol. 2020, 127. [Google Scholar] [CrossRef]
- Tian, Y.; Rao, H.; Tao, S.; Xue, W.T. Effect of boiling on the structure and immunoreactivity of recombinant peanut protein Ara h 1. Food Agric. Immunol. 2018, 29, 845–858. [Google Scholar] [CrossRef]
- Comstock, S.S.; Maleki, S.J.; Teuber, S.S.; Thierry, C. Boiling and Frying Peanuts Decreases Soluble Peanut (Arachis hypogaea) Allergens Ara h 1 and Ara h 2 But Does not Generate Hypoallergenic Peanuts. PLoS ONE 2016, 11, e0157849. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Rao, H.; Fu, W.; Tao, S.; Xue, W.-T. Effect of digestion on the immunoreactivity and proinflammatory properties of recombinant peanut allergen Ara h 1. Food Agric. Immunol. 2019, 30, 418–431. [Google Scholar] [CrossRef] [Green Version]
- Novak, N.; Maleki, S.J.; Cuadrado, C.; Crespo, J.F.; Cabanillas, B. Interaction of Monocyte-Derived Dendritic Cells with Ara h 2 from Raw and Roasted Peanuts. Foods 2020, 9, 863. [Google Scholar] [CrossRef]
- Kuraishi, C.; Nakagoshi, H.; Tanno, H.; Tanaka, H. Application of transglutaminase for food processing. In Hydrocolloids; Elsevier: Amsterdam, The Netherlands, 2000; pp. 281–285. [Google Scholar] [CrossRef]
- Chang, X.; Wu, Z.; Zhao, R.; Zhang, Y.; Li, X.; Yang, A.; Tong, P.; Chen, H. Analysis on MTGase catalysed cross-linked products of Ara h 2: Structure and immunoreactivity. Food Agric. Immunol. 2018, 29, 1197–1208. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Zhao, R.; Ren, L.; Li, X.; Yang, A.; Tong, P.; Chen, H. Modification of the reaction system of Ara h 2 catalyzed by MTGase: Products and reaction conditions analysis. J. Food Process. Preserv. 2017, 42, e13422. [Google Scholar] [CrossRef]
- Wu, Z.; Lian, J.; Zhao, R.; Li, K.; Li, X.; Yang, A.; Tong, P.; Chen, H. Ara h 2 cross-linking catalyzed by MTGase decreases its allergenicity. Food Funct. 2017, 8, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Tong, P.; Chen, S.; Gao, J.; Li, X.; Wu, Z.; Yang, A.; Yuan, J.; Chen, H. Caffeic acid-assisted cross-linking catalyzed by polyphenol oxidase decreases the allergenicity of ovalbumin in a Balb/c mouse model. Food Chem. Toxicol. 2017, 111, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Ma, J.; Ahmed, I.; Lv, L.; Li, Z.; Lin, H. Effect of tyrosinase-catalyzed crosslinking on the structure and allergenicity of turbot parvalbumin mediated by caffeic acid. J. Sci. Food Agric. 2019, 99, 3501–3508. [Google Scholar] [CrossRef]
- Ahmed, I.; Lv, L.; Lin, H.; Li, Z.; Ma, J.; Guanzhi, C.; Sun, L.; Xu, L. Effect of tyrosinase-aided crosslinking on the IgE binding potential and conformational structure of shrimp (Metapenaeus ensis) tropomyosin. Food Chem. 2017, 248, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Nagy, E.D.; Guo, Y.; Tang, S.; Bowers, J.E.; Okashah, R.A.; Taylor, C.A.; Zhang, D.; Khanal, S.; Heesacker, A.F.; Khalilian, N.; et al. A high-density genetic map of Arachis duranensis, a diploid ancestor of cultivated peanut. BMC Genom. 2012, 13, 469. [Google Scholar] [CrossRef] [Green Version]
- Kiewiet, M.B.; Dekkers, R.; Ulfman, L.H.; Groeneveld, A.; de Vos, P.; Fass, M.M. Immunomodulating protein aggregates in soy and whey hydrolysates and their resistance to digestion in an in vitro infant gastrointestinal model: New insights in the mechanism of immunomodulatory hydrolysates. Food Funct. 2018, 9, 604–613. [Google Scholar] [CrossRef]
- Tan, F.J.; Lai, K.M.; Hsu, K.C. A Comparative Study on Physical Properties and Chemical Interactions of Gels from Tilapia Meat Pastes induced by Heat and Pressure. J. Texture Stud. 2010, 41, 153–170. [Google Scholar] [CrossRef]
- Rao, H.; Tian, Y.; Tao, S.; Tang, J.; Li, X.; Xue, W.T. Key factors affecting the immunoreactivity of roasted and boiled peanuts: Temperature and water. LWT Food Sci. Technol. 2016, 72, 492–500. [Google Scholar] [CrossRef]
- Mondoulet, L.; Paty, E.; Drumare, M.F.; Ah-Leung, S.; Scheinmann, P.; Willemot, R.M.; Wal, J.M.; Bernard, H. Influence of thermal processing on the allergenicity of peanut proteins. J. Agric. Food Chem. 2005, 53, 4547–4553. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Rao, H.; Zhang, K.; Tao, S.; Xue, W.T. Effects of different thermal processing methods on the structure and allergenicity of peanut allergen Ara h 1. Food Sci. Nutr. 2018, 6, 1706–1714. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.pymol.org/ (accessed on 13 October 2020).
- Available online: http://www.rcsb.org/structure/3SMH (accessed on 13 October 2020).
- Chruszcz, M.; Maleki, S.J.; Majorek, K.A.; Demas, M.; Bublin, M.; Solberg, R.; Hurlburt, B.K.; Ruan, S.; Mattison, C.P.; Breiteneder, H.; et al. Structural and immunologic characterization of Ara h 1, a major peanut allergen. J. Biol. Chem. 2011, 286, 44294. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Tao, S.; Xue, J.; Zhang, H.; Xue, W.; Chen, F. Identification and purification of a novel fish allergen from largemouth bass (Micropterus salmoides). Food Agric. Immunol. 2014, 25, 70–81. [Google Scholar] [CrossRef]
- Hurlburt, B.K.; Mcbride, J.K.; Nesbit, J.B.; Ruan, S.; Maleki, S.J. Purification of Recombinant Peanut Allergen Ara h 1 and Comparison of IgE Binding to the Natural Protein. Foods 2014, 3, 642–657. [Google Scholar] [CrossRef] [Green Version]
- Cabanos, C.; Tandang-Silvas, M.R.; Odijk, V.; Brostedt, P.; Tanaka, A.; Utsumi, S.; Maruyama, N. Expression, purification, cross-reactivity and homology modeling of peanut profilin. Protein Expr. Purif. 2010, 73, 36–45. [Google Scholar] [CrossRef]
- Nielsen, P.M. Reactions and potential industrial applications of transglutaminase. Review of literature and patents. Food Biotechnol. 1995, 9, 119–156. [Google Scholar] [CrossRef]
- Miwa, N. Innovation in the food industry using microbial transglutaminase: Keys to success and future prospects. Anal. Biochem. 2020, 597, 113638. [Google Scholar] [CrossRef]
- Fangzhou, Y.; Ishfaq, A.; Liangtao, L.; Zhaojie, L.; Zhenxing, L.; Hong, L.; Hang, L.; Jinxia, Z.; Shenglan, T.; Jiaju, M. Impacts of glycation and transglutaminase-catalyzed glycosylation with glucosamine on the conformational structure and allergenicity of bovine β-lactoglobulin. Food Funct. 2018, 9, 3944–3955. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Liu, C.; Xue, W.; Wang, Z. Crosslinked Recombinant-Ara h 1 Catalyzed by Microbial Transglutaminase: Preparation, Structural Characterization and Allergic Assessment. Foods 2020, 9, 1508. https://doi.org/10.3390/foods9101508
Tian Y, Liu C, Xue W, Wang Z. Crosslinked Recombinant-Ara h 1 Catalyzed by Microbial Transglutaminase: Preparation, Structural Characterization and Allergic Assessment. Foods. 2020; 9(10):1508. https://doi.org/10.3390/foods9101508
Chicago/Turabian StyleTian, Yang, Chenglong Liu, Wentong Xue, and Zhongfu Wang. 2020. "Crosslinked Recombinant-Ara h 1 Catalyzed by Microbial Transglutaminase: Preparation, Structural Characterization and Allergic Assessment" Foods 9, no. 10: 1508. https://doi.org/10.3390/foods9101508
APA StyleTian, Y., Liu, C., Xue, W., & Wang, Z. (2020). Crosslinked Recombinant-Ara h 1 Catalyzed by Microbial Transglutaminase: Preparation, Structural Characterization and Allergic Assessment. Foods, 9(10), 1508. https://doi.org/10.3390/foods9101508