Removal of Antibiotics Using an Algae-Algae Consortium (Chlorella protothecoides and Chlorella vulgaris)
Abstract
:1. Introduction
2. Materials and Methods
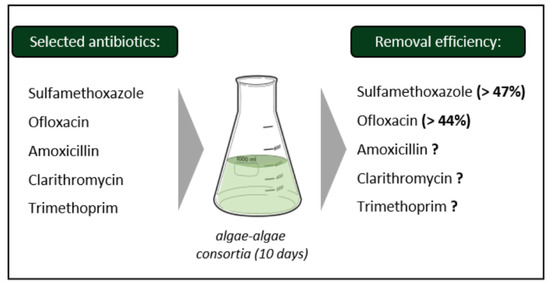
2.1. Selection of Target Antibiotics
2.2. Experimental Design
2.3. Algae Performance
2.4. Chlorophyll and Pigments Extraction and Quantification
2.5. Sample Preparation and Antibiotic SPE Extraction
2.6. Chemicals and Materials
2.7. Instrumental and Analytical Methodology
2.8. Quality Assurance Procedures
2.9. Data and Statistical Analyses
3. Results
3.1. Algae Performance
3.2. Analytical Validation
3.3. Antibiotic Removal Efficiency by the Algae Consortium
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lagadinou, M.; Onisor, M.O.; Rigas, A.; Musetescu, D.-V.; Gkentzi, D.; Assimakopoulos, S.F.; Panos, G.; Marangos, M. Antimicrobial Properties on Non-Antibiotic Drugs in the Era of Increased Bacterial Resistance. Antibiotics 2020, 9, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imperial, I.C.V.J.; Ibana, J.A. Addressing the Antibiotic Resistance Problem with Probiotics: Reducing the Risk of Its Double-Edged Sword Effect. Front. Microbiol. 2016, 7, 1983. [Google Scholar] [CrossRef] [Green Version]
- Dingmann, B.J. Searching for New Antibiotics Right Under our Feet. J. Public Health Issues Pract. 2018, 2, 111. [Google Scholar] [CrossRef] [Green Version]
- Pepi, M.; Focardi, S. Antibiotic-Resistant Bacteria in Aquaculture and Climate Change: A Challenge for Health in the Mediter-ranean Area. Int. J. Environ. Res. Public Health 2021, 18, 5723. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Li, Y.; Zhang, L.; Xiao, N. Environmental behavior and impacts of antibiotics. In Proceedings of the 6th International Conference on Energy, Environment and Sustainable Development (ICEESD 2017), Zhuhai, China, 11–12 March 2017; Atlantis Press: Amsterdam, The Netherlands, 2017; pp. 443–447. [Google Scholar]
- Bilal, M.; Mehmood, S.; Rasheed, T.; Iqbal, H.M. Antibiotics traces in the aquatic environment: Persistence and adverse envi-ronmental impact. Curr. Opin. Environ. Sci. Health 2020, 13, 68–74. [Google Scholar] [CrossRef]
- Kovalakova, P.; Leslie, C.; Thomas, J.M.; Blahoslav, M.; Mingbao, F.; Virender, K.S. Occurrence and toxicity of antibiotics in the aquatic environment: A review. Chemosphere 2020, 251, 26351. [Google Scholar] [CrossRef]
- Larsson, D.J. Antibiotics in the environment. Ups. J. Med. Sci. 2014, 119, 108–112. [Google Scholar] [CrossRef]
- Sabri, N.; van Holst, S.; Schmitt, H.; van der Zaan, B.; Gerritsen, H.; Rijnaarts, H.; Langenhoff, A. Fate of antibiotics and antibiotic resistance genes during conventional and additional treatment technologies in wastewater treatment plants. Sci. Total Environ. 2020, 741, 140199. [Google Scholar] [CrossRef]
- Phoon, B.L.; Ong, C.C.; Saheed, M.S.M.; Show, P.-L.; Chang, J.-S.; Ling, T.C.; Lam, S.S.; Juan, J.C. Conventional and emerging technologies for removal of antibiotics from wastewater. J. Hazard. Mater. 2020, 400, 122961. [Google Scholar] [CrossRef]
- Kang, Z.; Jia, X.; Zhang, Y.; Kang, X.; Ge, M.; Liu, D.; Wang, C.; He, Z. A Review on Application of Biochar in the Removal of Pharmaceutical Pollutants through Adsorption and Persulfate-Based AOPs. Sustainability 2022, 14, 10128. [Google Scholar] [CrossRef]
- Wang, C.; Sun, R.; Huang, R.; Wang, H. Superior fenton-like degradation of tetracycline by iron loaded graphitic carbon derived from microplastics: Synthesis, catalytic performance, and mechanism. Sep. Purif. Technol. 2021, 270, 118773. [Google Scholar] [CrossRef]
- Dusengemungu, L.; Kasali, G.; Gwanama, C.; Ouma, K.O. Recent Advances in Biosorption of Copper and Cobalt by Filamentous Fungi. Front. Microbiol. 2020, 11, 582016. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Show, P.L.; Ngo, H.H.; Ho, S.-H. Algae-mediated antibiotic wastewater treatment: A critical review. Environ. Sci. Ecotechnology 2022, 9, 100145. [Google Scholar] [CrossRef] [PubMed]
- Algae Biomass Organization. Algae in Advanced Wastewater Treatment. Available online: https://algaebiomass.org/blog/11590/algae-in-advanced-wastewater-treatment/ (accessed on 13 March 2023).
- Hejna, M.; Kapuścińska, D.; Aksmann, A. Pharmaceuticals in the Aquatic Environment: A Review on Eco-Toxicology and the Remediation Potential of Algae. Int. J. Environ. Res. Public Health 2022, 19, 7717. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Bhandari, G.; Bhatt, K.; Simsek, H. Microalgae-based removal of pollutants from wastewaters: Occurrence, toxicity and circular economy. Chemosphere 2022, 306, 135576. [Google Scholar] [CrossRef]
- Kerschgens, I.P.; Gademann, K. Antibiotic Algae by Chemical Surface Engineering. Chembiochem 2018, 19, 439–443. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Wang, J.; Li, H.; Mao, S.; Wang, D.; Xiang, Z.; Guo, R.; Chen, J. The dual function of the algal treatment: Antibiotic elimination combined with CO2 fixation. Chemosphere 2018, 211, 192–201. [Google Scholar] [CrossRef]
- Shchelik, I.S.; Sieber, S.; Gademann, K. Green Algae as a Drug Delivery System for the Controlled Release of Antibiotics. Chem. A Eur. J. 2020, 26, 16644–16648. [Google Scholar] [CrossRef]
- Shi, W.; Wang, L.; Rousseau, D.P.; Lens, P.N. Removal of estrone, 17α-ethinylestradiol, and 17ß-estradiol in algae and duckweed-based wastewater treatment systems. Environ. Sci. Poll. Res. 2010, 17, 824–833. [Google Scholar] [CrossRef]
- Xiong, J.-Q.; Kurade, M.B.; Jeon, B.-H. Ecotoxicological effects of enrofloxacin and its removal by monoculture of microalgal species and their consortium. Environ. Pollut. 2017, 226, 486–493. [Google Scholar] [CrossRef]
- Xiao, G.; Chen, J.; Show, P.L.; Yang, Q.; Ke, J.; Zhao, Q.; Guo, R.; Liu, Y. Evaluating the application of antibiotic treatment using algae-algae/activated sludge system. Chemosphere 2021, 282, 130966. [Google Scholar] [CrossRef]
- Matamoros, V.; Uggetti, E.; García, J.; Bayona, J.M. Assessment of the mechanisms involved in the removal of emerging contaminants by microalgae from wastewater: A laboratory scale study. J. Hazard. Mater. 2016, 301, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Michelon, W.; Matthiensen, A.; Viancelli, A.; Fongaro, G.; Gressler, V.; Soares, H.M. Removal of veterinary antibiotics in swine wastewater using microalgae-based process. Environ. Res. 2022, 207, 112192. [Google Scholar] [CrossRef]
- Leng, L.; Wei, L.; Xiong, Q.; Xu, S.; Li, W.; Lv, S.; Lu, Q.; Wan, L.; Wen, Z.; Zhou, W. Use of microalgae based technology for the removal of antibiotics from wastewater: A review. Chemosphere 2019, 238, 124680. [Google Scholar] [CrossRef]
- Morais, E.G.; Cristofoli, N.L.; Maia, I.B.; Magina, T.; Cerqueira, P.R.; Teixeira, M.R.; Varela, J.; Barreira, L.; Gouveia, L. Micro-algal systems for wastewater treatment: Technological trends and challenges towards waste recovery. Energies 2021, 14, 8112. [Google Scholar] [CrossRef]
- Guo, W.-Q.; Zheng, H.-S.; Li, S.; Du, J.-S.; Feng, X.-C.; Yin, R.-L.; Wu, Q.-L.; Ren, N.-Q.; Chang, J.-S. Removal of cephalosporin antibiotics 7-ACA from wastewater during the cultivation of lipid-accumulating microalgae. Bioresour. Technol. 2016, 221, 284–290. [Google Scholar] [CrossRef]
- Peng, F.-Q.; Ying, G.-G.; Yang, B.; Liu, S.; Lai, H.-J.; Liu, Y.-S.; Chen, Z.-F.; Zhou, G.-J. Biotransformation of progesterone and norgestrel by two freshwater microalgae (Scenedesmus obliquus and Chlorella pyrenoidosa): Transformation kinetics and products identification. Chemosphere 2014, 95, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Grey, D.; Sadoff, C.W. Sink or Swim? Water security for growth and development. Water Policy 2007, 9, 545–571. [Google Scholar] [CrossRef]
- Herbig, F.J. Talking dirty-effluent and sewage irreverence in South Africa: A conservation crime perspective. Cogent Soc. Sci. 2019, 5, 1701359. [Google Scholar] [CrossRef]
- de Jesus, J.H.; Lima, K.V.; Nogueira, R.F.P. Copper-containing magnetite supported on natural clay as a catalyst for heter-ogeneous photo-Fenton degradation of antibiotics in WWTP effluent. J. Environ. Chem. Eng. 2022, 10, 107765. [Google Scholar] [CrossRef]
- Brienza, M.; Sauvêtre, A.; Ait-Mouheb, N.; Bru-Adan, V.; Coviello, D.; Lequette, K.; Patureau, D.; Chiron, S.; Wéry, N. Reclaimed wastewater reuse in irrigation: Role of biofilms in the fate of antibiotics and spread of antimicrobial resistance. Water Res. 2022, 221, 118830. [Google Scholar] [CrossRef] [PubMed]
- Genthe, B.; Ndlela, L.; Madlala, A.T. Antimicrobial resistance screening and profiles: A glimpse from the South African per-spective. J. Water Health 2020, 18, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and Carotenoids: Measurement and Characterization by UV-VIS Spectroscopy. Curr. Protoc. Food Anal. Chem. 2001, 1, F4.3.1–F4.3.8. [Google Scholar] [CrossRef]
- ICH Harmonised Tripartite Guideline. Validation of Analytical Procedures: Text and Methodology. Q2 (R1); Somatek Inc.: San Diego, CA, USA, 2005. [Google Scholar]
- Oberholster, P.J.; Cheng, P.-H.; Genthe, B.; Steyn, M. The environmental feasibility of low-cost algae-based sewage treatment as a climate change adaption measure in rural areas of SADC countries. J. Appl. Phycol. 2018, 31, 355–363. [Google Scholar] [CrossRef]
- Elloum, W.; Jebali, A.; Maalej, A.; Chamkha, M.; Sayadi, S. Effect of Mild Salinity Stress on the Growth, Fatty Acid and Ca-rotenoid Compositions, and Biological Activities of the Thermal Freshwater Microalgae Scenedesmus sp. Biomolecules 2020, 10, 1515. [Google Scholar] [CrossRef]
- Patrolecco, L.; Rauseo, J.; Ademollo, N.; Grenni, P.; Cardoni, M.; Levantesi, C.; Luprano, M.; Caracciolo, A. Persistence of the antibiotic sulfamethoxazole in river water alone or in the co-presence of ciprofloxacin. Sci. Total Environ. 2018, 640, 1438–1446. [Google Scholar] [CrossRef]
- Zhong, X.; Zhu, Y.; Wang, Y.; Zhao, Q.; Huang, H. Effects of three antibiotics on growth and antioxidant response of Chlorella pyrenoidosa and Anabaena cylindrica Ecotoxicol. Environ. Saf. 2021, 211. [Google Scholar]
- Borecka, M.; Białk-Bielińska, A.; Haliński, P.; Pazdro, K.; Stepnowski, P.; Stolte, S. The influence of salinity on the toxicity of selected sulfonamides and trimethoprim towards the green algae Chlorella vulgaris. J. Hazard. Mater. 2016, 308, 179–186. [Google Scholar] [CrossRef]
- Zhang, Y.; Wan, J.; Li, Z.; Wu, Z.; Dang, C.; Fu, J. Enhanced removal efficiency of sulfamethoxazole by acclimated microalgae: Tolerant mechanism, and transformation products and pathways. Bioresour. Technol. 2021, 347, 126461. [Google Scholar] [CrossRef]
- Li, Z.; Li, S.; Li, T.; Gao, X.; Zhu, L. Physiological and transcriptomic responses of Chlorella sorokiniana to ciprofloxacin reveal molecular mechanisms for antibiotic removal. iScience 2022, 25, 104638. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.-Q.; Kurade, M.B.; Jeon, B.-H. Biodegradation of levofloxacin by an acclimated freshwater microalga, Chlorella vulgaris. Chem. Eng. J. 2017, 313, 1251–1257. [Google Scholar] [CrossRef]
- Berg, M.F.V.D.; Botha, A.M.; Bierman, A.; Oberholster, P. Assessing Domestic Wastewater Effluent with a Battery of Bioassays after Treatment with a Specific Consortium of Microalgae and Different Flocculation Methods. Water Air Soil Pollut. 2020, 231, 257. [Google Scholar] [CrossRef]





| Target Compounds | Retention Time (min) | Precursor ion | Quantifiable Product ions (m/z) | Collision Energy (eV) | Tube Lens (V) |
|---|---|---|---|---|---|
| (m/z) [M + H]+ | |||||
| Amoxicillin | 2.53 | 366.14 | 114.1*, 134.1 *, 208.0 *, 86.2 | 41 *, 21 *, 32 *, 41 | 80 |
| Ofloxacin | 4.87 | 361.85 | 318.1, 261.0 *, 221.0 | 19, 27 *, 36 | 100 |
| Ofloxacin-d3 | 4.97 | 365.17 | 261.1, | 20 | 100 |
| Sulfamethoxazole | 4.85 | 254.05 | 108.2, 92.1, 156.2 * | 19, 23, 16 * | 103 |
| Sulfamethoxazole-d4 | 4.95 | 258.17 | 160.1 | 23 | 100 |
| Trimethoprim | 4.45 | 291.14 | 230.04 *, 123.08, 261.02 * | 23*, 33, 25 * | 92 |
| Trimethoprim-d9 | 4.43 | 300.19 | 234 | 35 | 90 |
| Acridine-d9 | 5.24 | 189.27 | 159.1, 187.2 | 36 *, 34 | 96 |
| Clarithromycin | 6.55 | 748.48 | 590.2 *, 158.0 *, 558.2, 116.0 | 20 *, 27*, 23, 31 | 95 |
| Clarithromycin 13C-d3 | 6.59 | 752.48 | 161.9 | 35 | 90 |
| Target Compounds | Linearity Curve | LOD (µg/L) | LOQ (µg/L) | Accuracy (%) | Recoveries (%) |
|---|---|---|---|---|---|
| Amoxicillin | Y = −0.0014 + 0.0384 X, R2 = 0.9957 | 32.20 | 97.76 | 107.3 ± 5.1 | ND |
| Ofloxacin | Y = −0.018 + 3.8292 X, R2 = 0.9986 | 14.06 | 42.60 | 95.2 ± 0.5 | 105.1 ± 33.6 |
| Sulfamethoxazole | Y = 0.0034 + 0.9227 X, R2 = 0.9895 | 10.32 | 31.28 | 89.8 ± 4.4 | 118.6 ± 18.7 |
| Trimethoprim | Y = 0.0784 + 4.7033 X, R2 = 0.9906 | 3.53 | 10.70 | 93.3 ± 9.1 | 120.6 ± 33.4 |
| Clarithromycin | Y = 19.003 + 71.701 X, R2 = 0.9869 | 9.75 | 29.56 | 100 ± 0.2 | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ndlela, L.L.; Schroeder, P.; Genthe, B.; Cruzeiro, C. Removal of Antibiotics Using an Algae-Algae Consortium (Chlorella protothecoides and Chlorella vulgaris). Toxics 2023, 11, 588. https://doi.org/10.3390/toxics11070588
Ndlela LL, Schroeder P, Genthe B, Cruzeiro C. Removal of Antibiotics Using an Algae-Algae Consortium (Chlorella protothecoides and Chlorella vulgaris). Toxics. 2023; 11(7):588. https://doi.org/10.3390/toxics11070588
Chicago/Turabian StyleNdlela, Luyanda L., Peter Schroeder, Bettina Genthe, and Catarina Cruzeiro. 2023. "Removal of Antibiotics Using an Algae-Algae Consortium (Chlorella protothecoides and Chlorella vulgaris)" Toxics 11, no. 7: 588. https://doi.org/10.3390/toxics11070588
APA StyleNdlela, L. L., Schroeder, P., Genthe, B., & Cruzeiro, C. (2023). Removal of Antibiotics Using an Algae-Algae Consortium (Chlorella protothecoides and Chlorella vulgaris). Toxics, 11(7), 588. https://doi.org/10.3390/toxics11070588