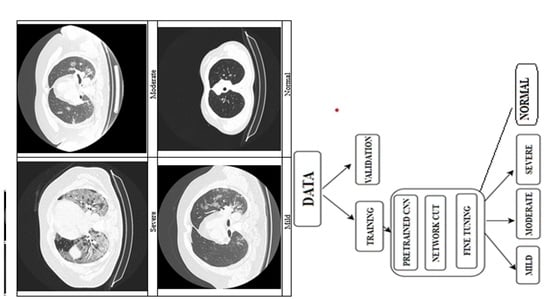
Classification of Pulmonary Damage Stages Caused by COVID-19 Disease from CT Scans via Transfer Learning
Abstract
:1. Introduction
- -
- -
- to differentiate the disease from common types of pneumonia: in [15], which uses pretrained networks such as VGG16 and RESNET50; in [16], which uses a multi-scale convolutional neural network with an area under the receiver operating characteristic curve (AUC) of 0.962; in [17], where normal cases were added, and Q-deformed entropy handcrafted features are classified using a long short-term memory network with a maximum accuracy of 99.68%;
- -
- to consider a data augmentation using a Fourier transform [18] for reducing the overfitting problems of lung segmentation.
2. Materials and Methods
2.1. Medical Context
2.2. Data Acquisition
2.3. Deep Learning Principles
2.4. Classification Algorithm Overview
3. Results
4. Discussion
- development of a new classification algorithm for the suggestive aspect of COVID-19 lung damage into mild, medium, or severe stages, for the axial view of computed tomography images using deep neural networks;
- enhancement of the five different online databases with new images collected from 55 patients;
- manual selection of the open lung phase images and their labeling into four classes.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AUC | Area under the receiver operating characteristic curve |
| CNN | Convolutional neural network |
| CT | Computerized tomography |
| DL | Deep learning |
| MCC | Matthew’s correlation coefficient |
| MERS-CoV | Middle East respiratory syndrome coronavirus |
| ML | Machine Learning |
| PCR | Polymerase Chain Reaction |
| SARS-CoV | Severe acute respiratory syndrome coronavirus |
| WHO | World Health Organization |
References
- Weiss, S.R.; Leibowitz, J.L. Coronavirus pathogenesis. Adv. Virus Res. 2011, 81, 85–164. [Google Scholar] [PubMed]
- Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 1 April 2021).
- Kooraki, S.; Hosseiny, M.; Myers, L.; Gholamrezanezhad, A. Coronavirus (COVID-19) Outbreak: What the Department of Radiology Should Know. J. Am. Coll. Radiol. 2020, 17, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Gietema, H.A.; Zelis, N.; Nobel, J.M.; Lambriks, L.J.G.; van Alphen, L.B.; Oude Lashof, A.M.L.; Wildberger, J.E.; Nelissen, I.C.; Stassen, P.M. CT in relation to RT-PCR in diagnosing COVID-19 in The Netherlands: A prospective study. PLoS ONE 2020, 15, e0235844. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Agarwal, P.P. Chest Imaging Appearance of COVID-19 Infection. Radiol. Cardiothorac. Imaging 2020, 2, e200028. [Google Scholar] [CrossRef] [Green Version]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A.; et al. CT Imaging Features of 2019 Novel Coronavirus (2019-nCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef] [Green Version]
- Santosh, K.C. AI-driven tools for coronavirus outbreak: Need of active learning and cross-population train/test models on multitudinal/ multimodal data. J. Med. Syst. 2020, 44, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Karthik, R.; Menaka, R.; Hariharan, M.; Won, D. Contour-enhanced attention CNN for CT-based COVID-19 segmentation. Pattern Recognit. 2022, 125, 108538. [Google Scholar] [CrossRef]
- Panwar, H.; Gupta, P.K.; Siddiqui, M.K.; Morales-Menendez, R.; Singh, V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos Solitons Fractals 2020, 138, 109944. [Google Scholar] [CrossRef]
- Namazi, H.; Kulish, V.V. Complexity-based classification of the coronavirus disease (COVID-19). Fractals 2020, 28, 2050114. [Google Scholar] [CrossRef]
- Available online: https://github.com/lindawangg/COVID-Net/ (accessed on 30 December 2020).
- Ahuja, S.; Panigrahi, B.K.; Dey, N.; Rajinikanth, V.; Gandhi, T.K. Deep transfer learning—Based automated detection of COVID-19 from lung CT scan slices. Appl. Intell. 2021, 51, 571–585. [Google Scholar] [CrossRef]
- Rahimzadeh, M.; Attar, A.; Sakhaei, S.M. A Fully Automated Deep Learning-based Network for Detecting COVID-19 from a New and Large Lung CT Scan Dataset. Biomed. Signal Process. Control 2021, 68, 102588. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Saba, L.; Gupta, S.K.; Carriero, A.; Falaschi, Z.; Paschè, A.; Danna, P.; El-Baz, A.; Naidu, S.; Suri, J.S. A Novel Block Imaging Technique Using Nine Artificial Intelligence Models for COVID-19 Disease Classification, Characterization and Severity Measurement in Lung Computed Tomography Scans on an Italian Cohort. J. Med. Syst. 2021, 45, 28. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.K.; Singh, P.; Joshi, S.D. Automated detection of COVID-19 from CT scan using convolutional neural network. Biocybern. Biomed. Eng. 2021, 41, 572–588. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Wong, P.K.; Ren, H.; Wang, H.; Wang, J.; Li, Y. Automatic distinction between COVID-19 and common pneumonia using multi-scale convolutional neural network on chest CT scans. Chaos Solitons Fractals 2020, 140, 110153. [Google Scholar] [CrossRef]
- Hasan, A.M.; L-Jawad, M.M.A.; Jalab, H.A.; Shaiba, H.; Ibrahim, R.W.; AL-Shamasneh, A.R. Classification of Covid-19 Coronavirus, Pneumonia and Healthy Lungs in CT Scans Using Q-Deformed Entropy and Deep Learning Features. Entropy 2020, 22, 517. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jiang, Y.; Ko, H.; Loew, M. A teacher-student framework with Fourier Transform augmentation for COVID-19 infection segmentation in CT images. Biomed. Signal Process Control 2023, 79, 104250. [Google Scholar] [CrossRef] [PubMed]
- Ramtohul, T.; Cabel, L.; Paoletti, X.; Chiche, L.; Moreau, P.; Noret, A.; Vuagnat, P.; Cherel, P.; Tardivon, A.; Cottu, P.; et al. Quantitative CT Extent of Lung Damage in COVID-19 Pneumonia Is an Independent Risk Factor for Inpatient Mortality in a Population of Cancer Patients: A Prospective Study. Front. Oncol. 2020, 10, 1560. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Omar, S.; Motawea, A.M.; Yasin, R. High-resolution CT features of COVID-19 pneumonia in confirmed cases. Egypt. J. Radiol. Nucl. Med. 2020, 51, 121. [Google Scholar] [CrossRef]
- Bernheim, A.; Mei, X.; Huang, M.; Yang, Y.; Fayad, Z.A.; Zhang, N.; Diao, K.; Lin, B.; Zhu, X.; Li, K.; et al. Chest CT Findings in Coronavirus Disease-19 (COVID-19): Relationship to Duration of Infection. Radiology 2020, 295, 200463. [Google Scholar] [CrossRef]
- Zhao, W.; Zhong, Z.; Xie, X.; Yu, Q.; Liu, J. Relation Between Chest CT Findings and Clinical Conditions of Coronavirus Disease (COVID-19) Pneumonia: A Multicenter Study. Am. J. Roentgenol. 2020, 214, 1072–1077. [Google Scholar] [CrossRef]
- Battaglia, P.W.; Hamrick, J.B.; Bapst, V.; Sanchez-Gonzalez, A.; Zambaldi, V.; Malinowski, M.; Tacchetti, A.; Raposo, D.; Santoro, A.; Faulkner, R.; et al. Relational inductive biases, deep learning, and graph networksz. arXiv 2018, arXiv:1806.01261. [Google Scholar]
- Fourcade, A.; Khonsari, R.H. Deep learning in medical image analysis: A third eye for doctors. J. Stomatol. Oral Maxillofac. Surg. 2019, 120, 279–288. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barshooi, A.H.; Amirkhani, A. A novel data augmentation based on Gabor filter and convolutional deep learning for improving the classification of COVID-19 chest X-Ray images. Biomed. Signal Process. Control 2022, 72, 103326. [Google Scholar] [CrossRef] [PubMed]
- Bottou, L. Online Algorithms and Stochastic Approximations, Online Learning and Neural Networks; Cambridge University Press: Cambridge, UK, 1998; ISBN 978-0-521-65263-6. [Google Scholar]
- Available online: https://www.kaggle.com/datasets/andrewmvd/covid19-ct-scans (accessed on 1 May 2022).
- Available online: https://www.kaggle.com/ahmedali2019/pneumonia-sample-xrays (accessed on 1 May 2022).
- Available online: https://www.kaggle.com/code/khoongweihao/covid-19-ct-scan-xray-cnn-detector (accessed on 1 May 2022).
- Available online: https://www.kaggle.com/datasets/luisblanche/covidct (accessed on 1 May 2022).
- Available online: https://www.kaggle.com/datasets/maedemaftouni/large-covid19-ct-slice-dataset (accessed on 1 May 2022).
- Roccetti, M.; Delnevo, G.; Casini, L.; Cappiello, G. Is bigger always better? A controversial journey to the center of machine learning design, with uses and misuses of big data for predicting water meter failures. J. Big Data 2019, 6, 70. [Google Scholar] [CrossRef] [Green Version]
- Roccetti, M.; Delnevo, G.; Casini, L.; Mirri, S. An alternative approach to dimension reduction for pareto distributed data: A case study. J. Big Data 2021, 8, 39. [Google Scholar] [CrossRef]








| Parameters | Unit | Values |
|---|---|---|
| Exposure Time | ms | 600 |
| Tube Current | mA | 106 |
| Color Type | - | grayscale |
| Bit Depth | - | 12 |
| Intensifier Size | mm | 250 |
| Image Width | pixels | 512 |
| Image Height | pixels | 512 |
| Number of frames per series | frames | 200–350 |
| No. | Hyperparameter | Value |
|---|---|---|
| 1 | Algorithm Type | Adam |
| 2 | No. of Epochs | 30 |
| 3 | Learn-Rate Schedule | piecewise |
| 4 | Learn-Rate Drop Factor | 0.2 |
| 5 | Learn-Rate Drop Period | 5 |
| 6 | Mini-Batch Size | 32 |
| 7 | Initial Learn Rate | 10−4 |
| 8 | Validation Frequency | 40 |
| 9 | Validation Patience | 15 |
| 10 | Shuffle | Every epoch |
| No. Patients/No. Images | Total Images | Mild | Moderate | Normal | Severe |
|---|---|---|---|---|---|
| Training | 4754 | 832 | 1021 | 2454 | 447 |
| Validation | 1188 | 208 | 255 | 613 | 112 |
| Testing | 1358 | 244 | 286 | 714 | 114 |
| Training Network | Accuracy (%) | Recall | Specificity | Precision | False Positive Rate | F1 Score | Matthews’ Correlation Coefficient | Cohen’s Kappa Coefficient |
|---|---|---|---|---|---|---|---|---|
| Resnet 50 | 86.89 (86.01,87.19) | 0.8 (0.79,0.82) | 0.96 (0.96,0.96) | 0.81 (0.78,0.81) | 0.04 (0.04,0.04) | 0.79 (0.78,0.8) | 0.76 (0.74,0.77) | 0.65 (0.63,0.66) |
| Inceptionv3 | 85.99 (82.11,86.97) | 0.78 (0.72,0.8) | 0.96 (0.95,0.96) | 0.8 (0.75,0.82) | 0.04 (0.04,0.05) | 0.77 (0.71,0.8) | 0.74 (0.68,0.77) | 0.63 (0.52,0.65) |
| Googlenet | 83.4 (82.4,84.54) | 0.76 (0.74,0.77) | 0.95 (0.95,0.95) | 0.78 (0.77,0.79) | 0.05 (0.05,0.05) | 0.74 (0.72,0.76) | 0.71 (0.69,0.73) | 0.56 (0.53,0.59) |
| Mobilenetv2 | 84.15 (83.65,86.75) | 0.76 (0.74,0.81) | 0.95 (0.95,0.96) | 0.77 (0.76,0.8) | 0.05 (0.04,0.05) | 0.75 (0.74,0.8) | 0.71 (0.7,0.76) | 0.58 (0.56,0.65) |
| Squeenet | 85.71 (84.17,87.78) | 0.79 (0.76,0.81) | 0.96 (0.95,0.96) | 0.79 (0.77,0.82) | 0.04 (0.04,0.05) | 0.78 (0.75,0.81) | 0.75 (0.72,0.78) | 0.62 (0.58,0.67) |
| Shufflenet | 80.65 (80.04,80.65) | 0.73 (0.71,0.73) | 0.94 (0.94,0.94) | 0.73 (0.72,0.73) | 0.06 (0.06,0.06) | 0.69 (0.68,0.69) | 0.66 (0.65,0.66) | 0.48 (0.47,0.48) |
| Class | Precision | Recall | F1 Score | False Positive Rate | Specificity |
|---|---|---|---|---|---|
| Mild (%) | 67.816 | 92.376 | 78.07 | 9.712 | 90.288 |
| Moderate (%) | 78.214 | 54.334 | 63.738 | 4.16 | 95.84 |
| Normal (%) | 99.972 | 100 | 99.986 | 0.032 | 99.968 |
| Severe (%) | 77.568 | 74.738 | 76.018 | 2.01 | 97.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tache, I.A.; Glotsos, D.; Stanciu, S.M. Classification of Pulmonary Damage Stages Caused by COVID-19 Disease from CT Scans via Transfer Learning. Bioengineering 2023, 10, 6. https://doi.org/10.3390/bioengineering10010006
Tache IA, Glotsos D, Stanciu SM. Classification of Pulmonary Damage Stages Caused by COVID-19 Disease from CT Scans via Transfer Learning. Bioengineering. 2023; 10(1):6. https://doi.org/10.3390/bioengineering10010006
Chicago/Turabian StyleTache, Irina Andra, Dimitrios Glotsos, and Silviu Marcel Stanciu. 2023. "Classification of Pulmonary Damage Stages Caused by COVID-19 Disease from CT Scans via Transfer Learning" Bioengineering 10, no. 1: 6. https://doi.org/10.3390/bioengineering10010006
APA StyleTache, I. A., Glotsos, D., & Stanciu, S. M. (2023). Classification of Pulmonary Damage Stages Caused by COVID-19 Disease from CT Scans via Transfer Learning. Bioengineering, 10(1), 6. https://doi.org/10.3390/bioengineering10010006