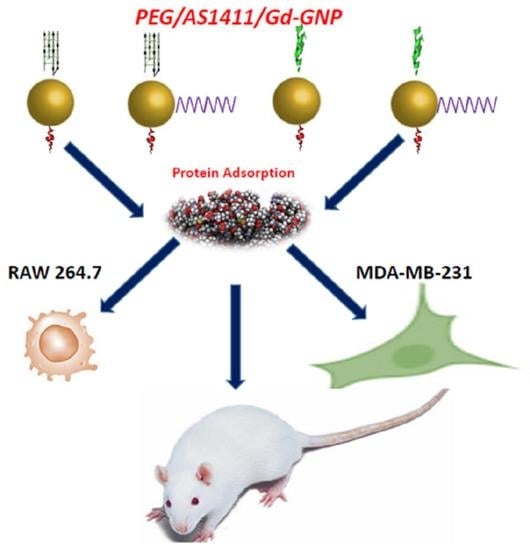
The Impact of PEGylation on Cellular Uptake and In Vivo Biodistribution of Gold Nanoparticle MRI Contrast Agents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. GNP Synthesis
2.3. Chemical Synthesis of Gadolinium (III) DO3A-SH (Gd)
2.4. Annealing and Preparing of AS1411 and CRO for Conjugation to GNPs
2.5. Preparation of PEG Solution
2.6. Conjugation of Gd(III)-DO3A-SH (Gd), AS1411 and/or CRO, and mPEG to GNP
2.7. Quantification of AS1411 and CRO per Nanoparticle
2.8. Quantification of Gadolinium
2.9. Protein Quantification by Bicinchoninic Acid (BCA) Assay
2.10. MDA-MB-231 and RAW 264.7 Cells Culture
2.11. Cellular Uptake
2.12. In Vivo Biodistribution Study
2.13. Statistical Analysis
3. Results
3.1. Synthesis and Characterization of GNPs
3.2. Number of Adsorbed Proteins on GNPs Surface
3.3. Cellular Uptake of GNPs
3.3.1. Uptake by Macrophage Cells
3.3.2. Uptake by Cancer Cells
3.4. In Vivo Biodistribution Study
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yeh, Y.; Creran, B.; Rotello, V. Gold nanoparticles: Preparation, properties, and applications in bionanotechnology. Nanoscale 2012, 4, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Freitas de Freitas, L.; Varca, G.; Batista, J.; Lugao, A. An overview of the synthesis of gold nanoparticles using radiation technologies. J. Nanomater. 2018, 8, 939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, Q.; Colon, J.; Cordero, A.; Bogdanovic, J.; Baker Ch Goodison, S.; Pensky, M. A facial nanoparticles immunoassay for cancer biomarker discovery. J. Nanobiotechnol. 2011, 23, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leuvering, J.; Thal, P.; Waart, M.; Schuurs, A. Sol particle immunoassay (SPIA). J Immun. 1980, 1, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Liang, X.; Deng, Z.; Feng Sh Li, X.; Huang, M.; Li, C.; Dai, Z. Prussian blue coated gold nanoparticles for simultaneous photoacoustic/CT bimodal imaging and photothermal ablation of cancer. Biomaterial 2014, 35, 5814–5821. [Google Scholar] [CrossRef]
- Wu, Y.; Ali, M.; Chen, K.; Fang, N.; El-Sayed, M. Gold nanoparticles in biological optical imaging. Nano Today 2019, 24, 120–140. [Google Scholar] [CrossRef]
- Wang, J.; O’Toole, M.G.; Massey, A.; Biswas, S.; Nantz, M.; Achilefu, S.; Kang, K.A. Highly specific, NIR fluorescent contrast agent with emission controlled by gold nanoparticle. In Oxygen Transport to Tissue XXXII; Elsevier: Amsterdam, The Netherlands, 2011; pp. 149–154. [Google Scholar]
- Vines, J.; Yoon, J.; Ryu, N.; Lim, D.; Park, H. Gold nanoparticles for photothermal cancer therapy. Front. Chem. 2019, 7, 167. [Google Scholar] [CrossRef] [Green Version]
- Jain, P.; Lee, K.; El-Sayed, I.; El-Sayed, M. Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: Applications in biological and biomedicine. J. Phys. Chem. B 2006, 110, 7238–7248. [Google Scholar] [CrossRef] [Green Version]
- Brann, T.; Patel, D.; Chauhan, R.; James, K.T.; Bates, P.J.; Malik, M.T.; Keynton, R.S.; O’Toole, M.G. Gold nanoplates as cancer-targeted photothermal actuators for drug delivery and triggered release. J. Nanomater. 2016, 2016, 11. [Google Scholar] [CrossRef]
- Sidiqque, S.; Chow, J. Gold nanoparticles for drug delivery and cancer therapy. Nanoscale 2020, 10, 3824. [Google Scholar] [CrossRef]
- Han, G.; Ghosh, P.; Rotello, V. Multi-functional gold nanoparticles for drug delivery. In Bio-Applications of Nanoparticles, 1st ed.; Chan, W.C.W., Ed.; Springer: New York, NY, USA, 2007; Volume 620, pp. 48–56. [Google Scholar]
- Zhang, Y.; Tan, J.; Zhou, L.; Shan, X.; Liu, J.; Ma, Y. Synthesis and application of AS1411-Functionalized gold nanoparticles for targeted therapy of gastric cancer. ACS Omega 2020, 23, 31233. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.; O’Toole, M.; Casson, L.; Thomas, S.; Bradi, G.; Reyes-Reyes, E.; Ng, C.; Kang, K.; Bates, P. AS1411- conjugates gold nanosphere and their potential for breast cancer therapy. Oncotarget 2015, 8, 22270–22281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Zhang, Y.; Wang, M.; Zhou, J.; Zhang, Q.; Yang, W.; Li, Y.; Yan, F. Gold nanoparticles combined human β-defensin 3 gene-modified human periodontal ligament cells alleviate periodontal destruction via the p38 MAPK pathway. Front Bioeng. Biotechnol. 2021, 28, 631191. [Google Scholar] [CrossRef] [PubMed]
- Suk, J.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2015, 99 Pt A, 28–51. [Google Scholar] [CrossRef] [Green Version]
- Chinen, A.; Guan, C.; Ko, C.; Mirkin, C. The impact of protein formation on the macrophage cellular uptake and biodistribution of spherical nucleic acids. Small 2017, 13, 1603847. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Meckes, B.; Mirkin, C. Spherical nucleic acids with tailored and active protein coronae. ACS Cent. Sci. 2019, 5, 1983–1990. [Google Scholar] [CrossRef] [Green Version]
- Akhter, M.; Khalilullah, H.; Gupta, M.; Alflaleh Alhakamy, N.; Riadi, Y.; Md, S. Impact of protein corona on the biological identity of nanomedicine; understanding the fate of nanomaterials in the biological milieu. Biomedicines 2021, 9, 1496. [Google Scholar] [CrossRef]
- Chen, F.; Wang, G.; Griffin, J.; Brenneman, B.; Banda, N.; Holers, V.; Backos, D.; Wu, L.; Moghimi, S.; Simberg, D. Complement proteins bind to nanoparticles protein corona and undergo dynamic exchange in vivo. Nat. Nanotechnol. 2017, 12, 387–393. [Google Scholar] [CrossRef] [Green Version]
- Tekie, F.; Hajiramezanali, M.; Geramifar, P.; Raoufi, M.; Dinarvand, R.; Soleimani, M.; Atyabi, F. Controlling evolution of protein corona: A prosperous approach to improve chitosan-based nanoparticles biodistribution and half-life. Sci. Rep. 2020, 10, 9664. [Google Scholar] [CrossRef]
- Lee, M.; Lee, H.; Rao, N.; Han, H.; Jeon, S.; Jeon, J.; Lee, S.; Kwon, S.; Suh, Y.; Park, J. Gold-stabilized carboxymethyl dextran nanoparticles for image-guided photodynamic therapy of cancer. J. Mater. Chem. B 2017, 21, 7319–7327. [Google Scholar] [CrossRef]
- Lee, K.; Chen, W.; Chen, Y. Using dextran-encapsulated gold nanoparticles as insulin carrier to prolong insulin activity. Nanomedicine 2017, 12, 1823–1834. [Google Scholar] [CrossRef] [PubMed]
- Bolanos, K.; Kogan, M.; Araya, E. Cappin gold nanoparticles with albumin to improve their biomedical properties. Int. J. Nanomed. 2019, 9, 6387–6406. [Google Scholar] [CrossRef] [Green Version]
- Fang, R.; Hu, C.h.; Zhang, L. Nanoparticles disguised as red blood cells to evade the immune system. Expert Opin. Biol. Ther. 2012, 12, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhang, Y.; Hou, X.; Feng, N. Red blood cell membrane-camouflaged nanoparticles: A novel drug delivery system for antitumor application. Acta Pharm. Sin. B 2019, 9, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Ma, G.; Ji, F.; Zhang, J.; Wang, L.; Sun, H.; Chen, S. Biocompatible long-circulating star carboxybetaine polymers. J. Mater. Chem. B 2015, 3, 440–448. [Google Scholar] [CrossRef]
- Li, D.; Wang, F.; Di, H.; Liu, X.; Zhang, P.; Zhou, W.; Liu, D. Cross-Linked poly (ethylene glycol) shells for nanoparticles: Enhanced stealth effect and colloidal stability. Langmuir 2019, 35, 8799–8805. [Google Scholar] [CrossRef] [PubMed]
- Quach, Q.; Kong, R.; Kah, J. Complement activation by PEGylated gold nanoparticles. Bioconjug. Chem. 2018, 29, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, B.; Pino, P.; Maffre, P.; Hartmann, R.; Gallego, M.; Rivera-Fernandez, S.; de la Fuente, J.; Nienhaus, G.; Parak, W. Surface functionalization of nanoparticles with polyethylene glycol: Effects on protein adsorption and cellular uptake. ACS Nano 2015, 9, 6996–7008. [Google Scholar] [CrossRef]
- Otsuka, H.; Nagasaki, Y.; Kataoka, K. PEGylated nanoparticles for biological and pharmaceutical applications. Adv. Drug Deliv. Rev. 2003, 24, 403–419. [Google Scholar] [CrossRef]
- Dos Santos, N.; Allen, C.; Doppen, A.M.; Anantha, M.; Cox, K.A.; Gallagher, R.C.; Karlsson, G.; Edwards, K.; Kenner, G.; Samuels, L.; et al. Influence of poly(ethylene glycol) grafting density and polymer length on liposomes: Relating plasma circulation lifetimes to protein binding. Biochim. Biophys. Acta 2007, 1768, 1367–1377. [Google Scholar] [CrossRef]
- Cruie, C.; Chithrani, D. Polyethylene glycol density and length affects nanoparticle uptake by cancer cells. J. Nanomed. Res. 2014, 1, 6. [Google Scholar]
- Shi, L.; Zhang, J.; Zhao, M.; Tang Sh Cheng, X.; Zhang, W.; Li, W.; Liu, X.; Peng, H.; Wang, Q. Effects of polyethylene glycol on the surface of nanoparticles for targeted drug delivery. Nanoscale 2021, 13, 10748–10764. [Google Scholar] [CrossRef] [PubMed]
- Walkey, C.; Olsen, J.; Guo, H.; Emili, A.; Chan, W. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 2012, 134, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, J.; Anchordoquy, T. Questioning the use of PEGylation for drug delivery. Drug Deliv. Transl. Res. 2013, 3, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Knop, K.; Hoogenboom, R.; Fischer, D.; Schubert, U.S. Poly(ethylene glycol) in drug delivery: Pros and cons as well as potential alternatives. Angew. Chem. 2010, 23, 6288–6308. [Google Scholar] [CrossRef]
- Abu Lila, A.; Kiwada, H.; Ishida, T. The accelerated blood clearance (ABC) phenomenon: Clinical challenge and approaches to manage. J. Control. Release 2013, 127, 38–47. [Google Scholar] [CrossRef]
- Saadati, R.; Dadashzadeh, S.; Abbasian, Z.; Soleimanjahi, H. Accelerated blood clearance of PEGylated PLGA nanoparticles following repeated injections: Effects of polymer dose, PEG coating, and encapsulated anticancer drug. Pharm. Res. 2013, 30, 985–995. [Google Scholar] [CrossRef]
- Chauhan, R.; El-Baz, N.; Keynton, R.; James, K.; Malik, D.; Zhu MEl-Baz, A.; Ng Ch Bates, P.; Malik, M.; O’Toole, M. Targeted gold nanoparticle–oligonucleotide contrast agents in combination with a new local voxel-wise MRI analysis algorithm for in vitro imaging of triple-negative breast cancer. J. Nanomater. 2019, 9, 709. [Google Scholar] [CrossRef] [Green Version]
- Bates, P.; Reyes-Reyes, E.; Malik, M.; Murphy, E.; O’Toole, M.; Trent, J. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta 2017, 1861, 1414–1428. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.; Salipur, F.; Shams, M.; Forsthoefel, M.; Bates, P. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol. Oncol. 2015, 9, 1392–1405. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Wang, Y.; Shi, H.; Dong, M.; Han, H.; Li, Q. Nucleolin-targeting AS1411 aptamer-modified micelle for the co-delivery of doxorubicin and miR-519c to improve the therapeutic efficacy in hepatocellular carcinoma treatment. Int. J. Nanomed. 2021, 16, 2569–2584. [Google Scholar] [CrossRef] [PubMed]
- Masitas, R.A.; Zamborini, F.P. Oxidation of highly unstable <4 nm diameter gold nanoparticles 850 mV negative of the bulk oxidation potential. J. Am. Chem. Soc. 2012, 134, 5014–5017. [Google Scholar] [PubMed]
- Haiss, W.; Thanh NT, K.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold nanoparticles from UV−Vis spectra. J. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef] [PubMed]
- Lebduskova, P.; Hermann, P.; Helm, L.; Toth, E.; Kotek, J.; Binnemans, K.; Rudovsky, J.; Lukes, I.; Merbach, A.E. Gadolinium(iii) complexes of mono- and diethyl esters of monophosphonic acid analogue of DOTA as potential MRI contrast agents: Solution structures and relaxometric studies. Dalton Trans. 2007, 4, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, S.; Campello, M.P.; Marques, F.; Gano, L.; Kubicek, V.; Fouskova, P.; Toth, E.; Santos, I. A novel tetraazamacrocycle bearing a thiol pendant arm for labeling biomolecules with radiolanthanides. Dalton Trans. 2009, 23, 4509–4518. [Google Scholar] [CrossRef]
- Chang, T.-C.; Yu, S.J. Microwave-assisted catalytic acetylation of alcohols by gold-nanoparticle–supported gadolinium complex. Synth. Commun. 2015, 45, 651–662. [Google Scholar] [CrossRef]
- Hurst, S.J.; Lytton-Jean, A.K.R.; Mirkin, C.A. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal. Chem. 2006, 78, 8313–8318. [Google Scholar] [CrossRef] [Green Version]
- Betker, J.; Gomez, J.; Anchordoquy, T. The effects of lipoplex formulation variables on the protein corona and comparisons with in vitro transfection efficiency. J. Control. Release 2013, 171, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Anchordoquy, T. Drug delivery trends in clinical trials and translational medicine: Challenges and opportunities in the delivery of nucleic acid-based therapeutics. J. Pharm. Sci. 2012, 100, 38–52. [Google Scholar] [CrossRef] [Green Version]
- Perera, Y.; Xu, J.; Amarasekara, D.; Hughes, A.; Abbood, I.; Fitzkee, N. Understanding the adsorption of peptides and proteins onto PEGylated gold nanoparticles. Molecules 2021, 26, 5788. [Google Scholar] [CrossRef]
- Li, M.; Jiang, S.; Simon, J.; Pablick, D.; Frey, M.; Wanger, M.; Mailander, V.; Crespy, D.; Landfester, K. Brush conformation of polyethylene glycol determine the stealth effect of nanocarriers in the low protein adsorption regime. Nano Lett. 2021, 21, 1591–1598. [Google Scholar] [CrossRef]
- Gref, R.; Luck, M.; Quellec, P.; Marchand, M.; Dellacherie, E.; Harnisch, S.; Blunk, T.; Muller, R.H. Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): Influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf. B. Biointerfaces 2000, 18, 301–313. [Google Scholar] [CrossRef]
- Hamad, I.; Al-Hanbali, O.; Hunter, A.C.; Rutt, K.J.; Andresen, T.L.; Moghimi, S.M. Distinct polymer architecture mediates switching of complement-activation pathways at the nanosphere-serum interface: Implications for stealth nanoparticle engineering. ACS Nano 2010, 4, 6629–6638. [Google Scholar] [CrossRef] [PubMed]
- Schaffler, M.; Semmler-Behnke, M.; Sarioglu, H.; Takenaka, S.; Wenk, A.; Schleh, C.; Hauck, S.; Johnston, B.; Kreyling, W. Serum protein identification and quantification of the corona of 5,15 and 80 nm gold nanoparticles. Nanotechnology 2013, 24, 265103. [Google Scholar] [CrossRef]
- Oh, N.; Park, J. Surface chemistry of gold nanoparticles mediates their exocytosis in macrophages. ACS Nano 2014, 8, 6232–6241. [Google Scholar] [CrossRef]
- Quach, Q.; Kah, J. Non-specific adsorption of complement proteins affects complement activation pathways of gold nanomaterials. Nanotoxicology 2017, 11, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Dezsi, L.; Fulop, T.; Meszaros, T.; Szenasi, G.; Urbanics, R.; Vazsonyi, C.; Orfi, E.; Rosivall Laszlo, L.; Nemes, R.; Kok, R.; et al. Features of complement activation-related pseudoallergy to liposomes with different surface charge and PEGylation: Comparison of the porcine and rat responses. J. Control. Release 2014, 195, 2–10. [Google Scholar] [CrossRef]
- Patel, P.; Giljohann, D.; Daniel, W.; Zheng, D.; Prigodich, A.; Mirkin, C. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjug. Chem. 2010, 21, 2250–2256. [Google Scholar] [CrossRef] [Green Version]
- Brun, E.; Sicard-Roselli, C. Could nanoparticle corona characterization help for biological consequence prediction? Cancer Nanotechnol. 2014, 5, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.; Tian, X.; Wu, A.; Li, J.; Tian, J.; Chong, Y.; Chai, Z.; Zhao, Y.; Chen, C.; Ge, C. Protein corona influences cellular uptake of gold nanoparticles by phagocytic and nonphagocytic cells in a size-dependent manner. ACS Appl. Mater. Interfaces 2015, 7, 20568–20575. [Google Scholar] [CrossRef]
- Mosquera, J.; Garcia, I.; Henriksen-Lacey, M.; Martinez-Calvo, M.; Dhanjani, M.; Mascarenas, J.; Liz-Marzan, L. Reversible control of protein corona formation on gold nanoparticles using host−guest interactions. ACS Nano 2020, 14, 5382–5391. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, A.; Salvati, A.; Santos-Martinez, M.; Radomski, M.; Dawson, K.; Aberg, C. Nanoparticle adhesion to the cell Membrane and Its effect on nanoparticle uptake efficiency. J. Am. Chem. Soc. 2013, 135, 1438–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosquera, J.; Gracia, I.; Liz-Marzan, L. Cellular uptake of nanoparticles versus small molecules: A matter of size. Acc. Chem. Res. 2018, 51, 2305–2313. [Google Scholar] [CrossRef] [Green Version]
- Petrarca, C.; Clemente, E.; Amato, V.; Pedata, P.; Sabbioni, E.; Bernardini, G.; Iavicoli, I.; Cortese, S.; Niu, Q.; Otsuki, T.; et al. Engineered metal-based nanoparticles and innate immunity. Clin. Mol. Allergy 2015, 13, 13. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.H.; Chang, L.W.; Lin, P. Metal-Based Nanoparticles and the Immune System: Activation, Inflammation and Potential Applications. Biomed. Res. Int. 2015, 2015, 143720. [Google Scholar] [CrossRef] [Green Version]
- Moyano, D.F.; Liu, Y.; Peer, D.; Rotello, V.M. Modulation of Immune Response Using Engineered Nanoparticle Surfaces. Small 2016, 12, 76–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Getts, D.R.; Shea, L.D.; Miller, S.D.; King, N.J.C. Harnessing nanoparticles for immune modulation. Trends Immunol. 2015, 36, 419–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.J.; Tietjen, G.T.; Saucier-sawyer, J.K.; Saltzman, W.M. A holistic approach to targeting disease with polymeric nanoparticles. Nat. Rev. Drug Disc. 2015, 14, 239–247. [Google Scholar] [CrossRef] [Green Version]
- Mohanan, D.; Slütter, B.; Henriksen-lacey, M.; Jiskoot, W.; Bouwstra, J.A.; Perrie, Y.; Kündig, T.M.; Gander, B.; Johansen, P. Administration routes affect the quality of immune responses: A cross-sectional evaluation of particulate antigen-delivery systems. J. Control. Release 2010, 147, 342–349. [Google Scholar] [CrossRef]








| GNP | Gd | AS1411/CRO | PEG |
|---|---|---|---|
| 4 nm non-PEGylated | 10× | 15× | - |
| 4 nm PEGylated | 6× | 10× | 6× |
| 10 nm non-PEGylated | 120× | 90× | - |
| 10 nm PEGylated | 72× | 60× | 36× |
| Sample | Oligonucleotide/ 4 nm GNPs | Oligonucleotide/ 10 nm GNPs | Gd/ 4 nm GNPs | Gd/ 10 nm GNPs |
|---|---|---|---|---|
| GNP-Gd-AS1411 | 10 | 43.2 | 6.5 | 77.2 |
| GNP-Gd-AS1411-PEG 1K | 7.4 | 43.7 | 5.7 | 64.7 |
| GNP-Gd-AS1411-PEG 2K | 7.6 | 46.8 | 5.9 | 55.1 |
| GNP-Gd-AS1411-PEG 5K | 7 | 31.5 | 6.5 | 72.6 |
| GNP-Gd-CRO | 12.3 | 44.9 | 9.0 | 150.0 |
| GNP-Gd-CRO-PEG 1K | 6.3 | 42.9 | 7.9 | 81.8 |
| GNP-Gd-CRO-PEG 2K | 8.3 | 38.9 | 6.4 | 115.2 |
| GNP-Gd-CRO-PEG 5K | 8.7 | 40.7 | 7.9 | 113.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Baz, N.; Nunn, B.M.; Bates, P.J.; O’Toole, M.G. The Impact of PEGylation on Cellular Uptake and In Vivo Biodistribution of Gold Nanoparticle MRI Contrast Agents. Bioengineering 2022, 9, 766. https://doi.org/10.3390/bioengineering9120766
El-Baz N, Nunn BM, Bates PJ, O’Toole MG. The Impact of PEGylation on Cellular Uptake and In Vivo Biodistribution of Gold Nanoparticle MRI Contrast Agents. Bioengineering. 2022; 9(12):766. https://doi.org/10.3390/bioengineering9120766
Chicago/Turabian StyleEl-Baz, Nagwa, Betty M. Nunn, Paula J. Bates, and Martin G. O’Toole. 2022. "The Impact of PEGylation on Cellular Uptake and In Vivo Biodistribution of Gold Nanoparticle MRI Contrast Agents" Bioengineering 9, no. 12: 766. https://doi.org/10.3390/bioengineering9120766
APA StyleEl-Baz, N., Nunn, B. M., Bates, P. J., & O’Toole, M. G. (2022). The Impact of PEGylation on Cellular Uptake and In Vivo Biodistribution of Gold Nanoparticle MRI Contrast Agents. Bioengineering, 9(12), 766. https://doi.org/10.3390/bioengineering9120766