SNiPhunter: A SNP-Based Search Engine
Abstract
:1. Summary
2. Data Description
3. Use Case
4. Methods
4.1. Parsing Semi-Structured Biomedical Literature
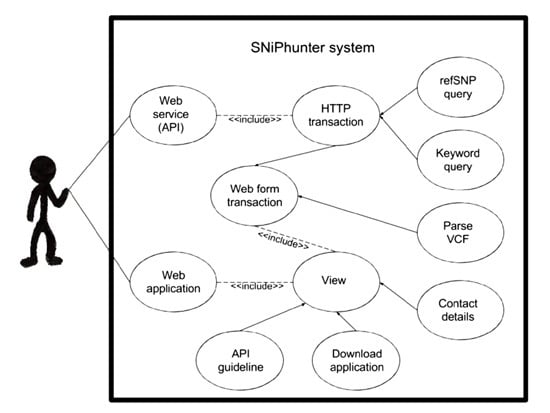
4.2. Implementing a Web Application and Web Service
4.3. Implementing a Database Interaction Protocol
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| API | Application programming interface |
| SNP | Single nucleotide polymorphism |
| NoSQL | Not only structured query language |
| SQL | Structured query language |
| VCF | Variant call format |
| refSNP | Reference single nucleotide polymorphism |
| PMC-OAI | Pubmed Central open access initiative |
| F/oss | Free and/or open source software |
| REST | Representational state transfer |
| XML | Extensible markup language |
| JSON | JavaScript object notation |
| HTML | Hypertext markup language |
| CSS | Cascading style sheets |
| AJAX | Asynchronous JavaScript and XML |
| DOM | Document object model |
References
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Machado, C.M.; Rebholz-Schuhmann, D.; Freitas, A.T.; Couto, F.M. The semantic web in translational medicine: Current applications and future directions. Brief. Bioinform. 2015, 16, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Price, W.N. Describing Black-Box Medicine. B.U. J. Sci. Tech. L. 2015, 21, 347. [Google Scholar]
- Zhang, J.; Baran, J.; Cros, A.; Guberman, J.M.; Haider, S.; Hsu, J.; Liang, Y.; Rivkin, E.; Wang, J.; Whitty, B.; et al. International Cancer Genome Consortium Data Portal—A one-stop shop for cancer genomics data. Database 2011. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Cariaso, M.; Lennon, G. SNPedia: A wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res. 2012, 40, D1308–D1312. [Google Scholar] [CrossRef] [PubMed]
- SNiPhunter. 2016. Available online: http://sniphunter.sanbi.ac.za (accessed on 27 September 2016).
- Pubmed. 2016. Available online: http://www.ncbi.nlm.nih.gov/pubmed (accessed on 27 September 2016).
- Google form. 2016. Available online: https://goo.gl/ccGgwb (accessed on 27 September 2016).
- Pubmed Central open access initiative. 2016. Available online: https://www.ncbi.nlm.nih.gov/pmc/tools/ftp/ (accessed on 27 September 2016).
- Directory of Open Access Journals. 2016. Available online: http://doaj.org (accessed on 27 September 2016).
- OpenDOAR. 2016. Available online: http://www.opendoar.org (accessed on 27 September 2016).
- CiteSeerX. 2016. Available online: http://citeseerx.ist.psu.edu/index (accessed on 27 September 2016).
- Shah, P.K.; Perez-Iratxeta, C.; Bork, P.; Andrade, M.A. Information extraction from full text scientific articles: Where are the keywords? BMC Bioinform. 2003, 4, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Baumgartner, W.A.; Randolph, D.A.; Cohen, K.B.; Hunter, L. MutationFinder: A high-performance system for extracting point mutation mentions from text. Bioinformatics 2007, 23, 1862–1865. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.H.; Harris, B.R.; Kao, H.Y.; Lu, Z. tmVar: A text mining approach for extracting sequence variants in biomedical literature. Bioinformatics 2013, 29, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Typicode. JSON-Server. 2015. Available online: https://github.com/typicode/json-server (accessed on 30 September 2015).
- Verou, L. Awesomplete. 2015. Available online: https://github.com/LeaVerou/awesomplete (accessed on 30 September 2015).
- Yu, G. Awk. 2015. Available online: https://github.com/guo-yu/awk (accessed on 14 December 2015).
- Robins, C. Forever. 2015. Available online: https://github.com/foreverjs/forever (accessed on 30 September 2015).
- Zapty Inc.; Agarwal, A. Forever-Service. 2015. Available online: https://github.com/zapty/forever-service (accessed on 30 September 2015).
- Sagalaev, I. Highlight. 2015. Available online: https://github.com/isagalaev/highlight.js (accessed on 30 November 2015).
- Farthin, S. Intercept-Stdout. 2015. Available online: https://github.com/sfarthin/intercept-stdout (accessed on 14 December 2015).
- Yaapa, H. Multer. 2015. Available online: https://github.com/expressjs/multer (accessed on 14 December 2015).
- Typicode. Lowdb. 2015. Available online: https://github.com/typicode/lowdb (accessed on 30 September 2015).




© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veldsman, W.P.; Christoffels, A. SNiPhunter: A SNP-Based Search Engine. Data 2016, 1, 17. https://doi.org/10.3390/data1030017
Veldsman WP, Christoffels A. SNiPhunter: A SNP-Based Search Engine. Data. 2016; 1(3):17. https://doi.org/10.3390/data1030017
Chicago/Turabian StyleVeldsman, Werner P., and Alan Christoffels. 2016. "SNiPhunter: A SNP-Based Search Engine" Data 1, no. 3: 17. https://doi.org/10.3390/data1030017
APA StyleVeldsman, W. P., & Christoffels, A. (2016). SNiPhunter: A SNP-Based Search Engine. Data, 1(3), 17. https://doi.org/10.3390/data1030017