Phylogenetic and Chemotaxonomic Studies Confirm the Affinities of Stromatoneurospora phoenix to the Coprophilous Xylariaceae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Survey and Sample Collection
2.2. Morphological Characterization
2.3. DNA Extraction, Polymerase Chain Reaction (PCR), and Sequencing
2.4. Phylogenetic Analyses
2.5. Analytical Equipment for Structure Elucidation
2.6. Fungal Material and Cultivation
2.7. Extraction and Isolation of Secondary Metabolites
2.8. Antimicrobial and Cytotoxic Activity Assay
2.9. Spectral Data
2.9.1. Phoenixilane A (1)
2.9.2. Phoenixilane B (2)
3. Results
3.1. Taxonomic and Phylogenetic Characterization
3.2. Molecular Phylogeny
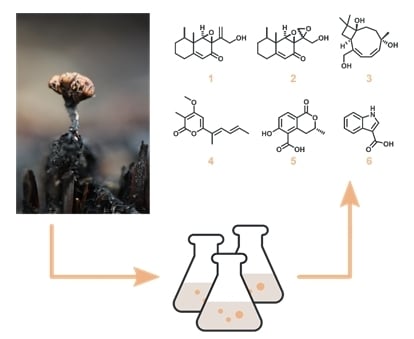
3.3. Isolation and Structure Elucidation of Secondary Metabolites
3.4. Biological Activities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jong, S.C.; Davis, E.E. Stromatic Neurosporas. Mycologia 1973, 65, 458–464. [Google Scholar] [CrossRef]
- Fries, E.M. Eclogae fungorum, praecipue ex herbarus germanorum de scriptorum. Linnaea 1830, 5, 497–553. [Google Scholar]
- Daranagama, D.A.; Hyde, K.D.; Sir, E.B.; Thambugala, K.M.; Tian, Q.; Samarakoon, M.C.; McKenzie, E.H.C.; Jayasiri, S.C.; Tibpromma, S.; Bhat, J.D.; et al. Towards a natural classification and backbone tree for Graphostromataceae, Hypoxylaceae, Lopadostomataceae and Xylariaceae. Fungal Divers. 2017, 88, 1–165. [Google Scholar] [CrossRef]
- Wendt, L.; Sir, E.B.; Kuhnert, E.; Heitkämper, S.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Luangsa-ard, J.J.; Srikitikulchai, P.; Peršoh, D.; et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2018, 17, 115–154. [Google Scholar] [CrossRef] [Green Version]
- Peláez, F.; González, V.; Platas, G.; Sánchez Ballesteros, J.; Rubio, V. Molecular phylogenetic studies within the Xylariaceae based on ribosomal DNA sequences. Fungal Divers. 2008, 31, 111–134. [Google Scholar]
- Jayawardena, R.S.; Purahong, W.; Zhang, W.; Wubet, T.; Li, X.; Liu, M.; Zhao, W.; Hyde, K.D.; Liu, J.; Yan, J. Biodiversity of fungi on Vitis vinifera L. revealed by traditional and high-resolution culture-independent approaches. Fungal Divers. 2018, 90, 1–84. [Google Scholar] [CrossRef] [Green Version]
- Wibberg, D.; Stadler, M.; Lambert, C.; Bunk, B.; Spröer, C.; Rückert, C.; Kalinowski, J.; Cox, R.J.; Kuhnert, E. High quality genome sequences of thirteen Hypoxylaceae (Ascomycota) strengthen the phylogenetic family backbone and enable the discovery of new taxa. Fungal Divers. 2020. [Google Scholar] [CrossRef]
- Sir, E.B.; Kuhnert, E.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Stadler, M. New species and reports of Hypoxylon from Argentina recognized by a polyphasic approach. Mycol. Prog. 2016, 15. [Google Scholar] [CrossRef]
- Kuhnert, E.; Sir, E.B.; Lambert, C.; Hyde, K.D.; Hladki, A.I.; Romero, A.I.; Rohde, M.; Stadler, M. Phylogenetic and chemotaxonomic resolution of the genus Annulohypoxylon (Xylariaceae) including four new species. Fungal Divers. 2017, 85, 1–43. [Google Scholar] [CrossRef]
- Rayner, R.W. A Mycological Colour Chart; Commonwealth Mycological Institute (Great Britain) Kew Gardens: Surrey, UK, 1970. [Google Scholar]
- Mackill, D.J. Classifying Japonica Rice Cultivars with RAPD Markers. Crop Sci. 1995, 35, 889–894. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunyard, B.A.; Nicholson, M.S.; Royse, D.J. A systematic assessment of Morchella using RFLP analysis of the 28S ribosomal RNA gene. Mycologia 1994, 86, 762–772. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 2013, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Nylander, J. MrModeltest V2; Uppsala Universitit: Uppsala, Sweden, 2004. [Google Scholar]
- Hsieh, H.M.; Ju, Y.M.; Rogers, J.D. Molecular phylogeny of Hypoxylon and closely related genera. Mycologia 2005, 97, 844–865. [Google Scholar] [CrossRef]
- Li, Q.R.; Kang, J.C.; Hyde, K.D. Two new species of the genus Collodiscula (Xylariaceae) from China. Mycol. Prog. 2015, 14. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Voglmayr, H. Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Divers. 2011, 52, 75–98. [Google Scholar] [CrossRef]
- Triebel, D.; Peršoh, D.; Wollweber, H.; Stadler, M. Phylogenetic relationships among Daldinia, Entonaema, and Hypoxylon as inferred from ITS nrDNA analyses of Xylariales. Nova Hedw. 2005, 80, 25–43. [Google Scholar] [CrossRef]
- Kuhnert, E.; Fournier, J.; Peršoh, D.; Luangsa-ard, J.J.D.; Stadler, M. New Hypoxylon species from Martinique and new evidence on the molecular phylogeny of Hypoxylon based on ITS rDNA and β-tubulin data. Fungal Divers. 2014, 64, 181–203. [Google Scholar] [CrossRef]
- Stadler, M.; Laessoe, T.; Fournier, J.; Decock, C.; Schmieschek, B.; Tichy, H.V.; Persoh, D. A polyphasic taxonomy of Daldinia (Xylariaceae). Stud. Mycol. 2014, 77, 1–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Castlebury, L.A.; Miller, A.N.; Huhndorf, S.M.; Schoch, C.L.; Seifert, K.A.; Rossman, A.Y.; Rogers, J.D.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B.; et al. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 2017, 98, 1076–1087. [Google Scholar] [CrossRef]
- Koukol, O.; Kelnarová, I.; Černý, K.; Woodward, S. Recent observations of sooty bark disease of sycamore maple in Prague (Czech Republic) and the phylogenetic placement of Cryptostroma corticale. For. Pathol. 2015, 45, 21–27. [Google Scholar] [CrossRef]
- Stadler, M.; Kuhnert, E.; Peršoh, D.; Fournier, J. The Xylariaceae as model example for a unified nomenclature following the “One Fungus-One Name” (1F1N) concept. Mycology 2013, 4, 5–21. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, J.D.; Jones, E.B.G.; McKenzie, E.H.C.; Dai, D.Q.; Daranagama, D.A.; Dayarathne, M.C.; Goonasekara, I.D.; et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 2015, 73, 73–144. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Bitzer, J.; Laessoe, T.; Fournier, J.; Kummer, V.; Decock, C.; Tichy, H.V.; Piepenbring, M.; Persoh, D.; Stadler, M. Affinities of Phylacia and the daldinoid Xylariaceae, inferred from chemotypes of cultures and ribosomal DNA sequences. Mycol. Res. 2008, 112, 251–270. [Google Scholar] [CrossRef]
- Serrano, R.; Gonzalez-Menendez, V.; Rodriguez, L.; Martin, J.; Tormo, J.R.; Genilloud, O. Co-culturing of fungal strains against Botrytis cinerea as a model for the induction of chemical diversity and therapeutic agents. Front. Microbiol. 2017, 8, 649. [Google Scholar] [CrossRef] [PubMed]
- Peláez, F.; Cabello, A.; Platas, G.; Díez, M.T.; del Val, A.G.; Basilio, A.; Martán, I.; Vicente, F.; Bills, G.F.; Giacobbe, R.A.; et al. The discovery of enfumafungin, a aovel antifungal compound produced by an Endophytic Hormonema species biological activity and taxonomy of the producing organisms. Syst. App. Microbiol. 2000, 23, 333–343. [Google Scholar] [CrossRef]
- Becker, K.; Wessel, A.-C.; Luangsa-ard, J.J.; Stadler, M. Viridistratins A−C, antimicrobial and cytotoxic benzo[j]fluoranthenes from stromata of Annulohypoxylon viridistratum (Hypoxylaceae, Ascomycota). Biomolecules 2020, 10, 805. [Google Scholar] [CrossRef] [PubMed]
- Sandargo, B.; Michehl, M.; Praditya, D.; Steinmann, E.; Stadler, M.; Surup, F. Antiviral meroterpenoid rhodatin and sesquiterpenoids rhodocoranes A-E from the Wrinkled Peach Mushroom, Rhodotus palmatus. Org. Lett. 2019, 21, 3286–3289. [Google Scholar] [CrossRef]
- Stadler, M.; Lambert, C.; Wibberg, D.; Kalinowski, J.; Cox, R.J.; Kolařík, M.; Kuhnert, E. Intragenomic polymorphisms in the ITS region of high-quality genomes of the Hypoxylaceae (Xylariales, Ascomycota). Mycol. Prog. 2020, 19, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.R.; Edwards, R.L.; Freer, A.A.; Mabelis, R.P.; Poyser, J.P.; Spencer, H.; Whalley, A.J.S. Punctatins B and C (antibiotics M95154 and M95155): Further sesquiterpene alcohols from the fungus Poronia punctata. J. Chem. Soc, Chem. Commun. 1984. [Google Scholar] [CrossRef]
- Schuffler, A.; Sterner, O.; Anke, H. Cytotoxic a-pyrones from Xylaria hypoxylon. Z. Nat. C J. Biosci. 2007, 62, 169–172. [Google Scholar] [CrossRef]
- Anderson, J.R.; Edwards, R.L.; Whalley, A.J.S. Metabolites of the higher fungi. Part 21. 3-Methyl-3,4-dihydroisocoumarins and related compounds from the ascomycete family Xylariaceae. J. Chem. Soc. Perkin Trans. 1 1983. [Google Scholar] [CrossRef]
- Fan, N.-W.; Chang, H.-S.; Cheng, M.-J.; Hsieh, S.-Y.; Liu, T.-W.; Yuan, G.-F.; Chen, I.-S. Secondary Metabolites from the Endophytic Fungus Xylaria cubensis. Helv. Chim. Acta 2014, 97, 1689–1699. [Google Scholar] [CrossRef]
- Subramanian, C.V.; Chandrashekara, K.V. Lindquistia indica new genus new species of hyphomycete. Bol. Soc. Argent. Bot. 1977, 18, 145–151. [Google Scholar]
- Rogers, J.D. Anamorphs of Xylaria: Taxonomic Considerations. Sydowia 1985, 38, 255–262. [Google Scholar]
- Márquez, S.S.; Bills, G.F.; Herrero, N.; Zabalgogeazcoa, Í. Non-systemic fungal endophytes of grasses. Fungal Ecol. 2012, 5, 289–297. [Google Scholar] [CrossRef]
- Kruys, Å.; Huhndorf, S.M.; Miller, A.N. Coprophilous contributions to the phylogeny of Lasiosphaeriaceae and allied taxa within Sordariales (Ascomycota, Fungi). Fungal Divers. 2014, 70, 101–113. [Google Scholar] [CrossRef]
- U’Ren, J.M.; Miadlikowska, J.; Zimmerman, N.B.; Lutzoni, F.; Stajich, J.E.; Arnold, A.E. Contributions of North American endophytes to the phylogeny, ecology, and taxonomy of Xylariaceae (Sordariomycetes, Ascomycota). Mol. Phylogenet. Evol. 2016, 98, 210–232. [Google Scholar] [CrossRef] [Green Version]
- Petrini, O. Taxonomy of endophytic fungi of aerial plant tissues. In Microbiology of the Phyllosphere; Fokkema, N.J., van den Heuvel, J., Eds.; Cambride University Press: Cambridge, UK, 1986; pp. 175–187. [Google Scholar]
- Zak, J.C.; Wicklow, D.T. Response of carbonicolous ascomycetes to aerated steam temperatures and treatment intervals. Can. J. Bot. 1978, 56, 2313–2318. [Google Scholar] [CrossRef]
- Carroll, G. Fungal Endophytes in Stems and Leaves: From Latent Pathogen to Mutualistic Symbiont. Ecology 1988, 69, 2–9. [Google Scholar] [CrossRef]
- Horie, Y.; Li, D. Five interesting Ascomycetes from herbal drugs. Mycoscience 1997, 38, 287–295. [Google Scholar] [CrossRef]
- Fisher, P.J.; Anson, A.E.; Petrini, O. Fungal endophytes in Ulex europaeus and Ulex gallii. Trans. Br. Mycol. Soc. 1986, 86, 153–156. [Google Scholar] [CrossRef]
- Stadler, M.; Ju, Y.M.; Rogers, J.D. Chemotaxonomy of Entonaema, Rhopalostroma and other Xylariaceae. Mycol. Res. 2004, 108, 239–256. [Google Scholar] [CrossRef]
- GBIF.org. GBIF Occurrence Download. Available online: https://doi.org/10.15468/dl.g84kjk (accessed on 22 June 2020).
- Rogers, J.D.; Ju, Y.M.; Hemmes, D.E. Hypoxylon rectangulosporum sp. nov., Xylaria psidii sp. nov., and Comments on Taxa of Podosordaria and Stromatoneurospora. Mycologia 2018, 84, 166–172. [Google Scholar] [CrossRef]
- Liang, Y.; Xu, W.; Liu, C.; Zhou, D.; Liu, X.; Qin, Y.; Cao, F.; Li, J.; Yang, R.; Qin, J. Eremophilane sesquiterpenes from the endophytic fungus Xylaria sp. GDG-102. Nat. Prod. Res. 2019, 33, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C.; Hsiao, G.; Lin, R.K.; Kuo, Y.H.; Ju, Y.M.; Lee, T.H. Bioactive constituents from the termite nest-derived medicinal fungus Xylaria nigripes. J. Nat. Prod. 2017, 80, 38–44. [Google Scholar] [CrossRef]
- Smith, C.J.; Morin, N.R.; Bills, G.F.; Dombrowski, A.W.; Salituro, G.M.; Smith, S.K.; Zhao, A.; MacNeil, D.J. Novel sesquiterpenoids from the fermentation of Xylaria persicaria are selective ligands for the NPY Y5 receptor. J. Org. Chem. 2002, 67, 5001–5004. [Google Scholar] [CrossRef]
- Ridderbusch, D.C.; Weber, R.W.; Anke, T.; Sterner, O. Tulasnein and podospirone from the coprophilous xylariaceous fungus Podosordaria tulasnei. Z. Nat. C 2004, 59, 379–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedras, M.S.; Zheng, Q.A.; Strelkov, S. Metabolic changes in roots of the oilseed canola infected with the biotroph Plasmodiophora brassicae: Phytoalexins and phytoanticipins. J. Agric. Food Chem. 2008, 56, 9949–9961. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.D. Sarcoxylon and Entonaema (Xylariaceae). Mycologia 2018, 73, 28–61. [Google Scholar] [CrossRef]






| Taxon | Strain/Status | Country | GenBank Accession Numbers | Reference | |||
|---|---|---|---|---|---|---|---|
| ITS | LSU | RPB2 | TUB2 | ||||
| Amphirosellinia fushanensis | HAST91111209/HT | Taiwan | GU339496 | N/A | GQ848339 | GQ495950 | [23] |
| Amph. nigrospora | HAST 91092308/HT | Taiwan | GU322457 | N/A | GQ848340 | GQ495951 | [23] |
| Anthostomella helicofissa | MFLUCC14-0173 | Italy | KP297406 | N/A | KP340534 | KP406617 | [3] |
| Anth. rubicola | MFLUCC16-0479 | Italy | KX533455 | KX533456 | N/A | N/A | [3] |
| Astrocystis concavispora | MFLUCC140.74 | Italy | KP297404 | KP340545 | KP340532 | KP406615 | [3] |
| Biscogniauxia nummularia | MUCL 51395/ET | France | KY610382 | KY610427 | KY624236 | KX271241 | [4] |
| Collodiscula bambusae | GZU H0102 | China | KP054279 | KP054280 | KP276675 | KP276674 | [24] |
| C. fangjingshanensis | GZU H0109/ET | China | KR002590 | KR002591 | KR002592 | KR002589 | [24] |
| C. japonica | CBS124266 | China | JF440974 | JF440974 | KY624273 | KY624316 | ITS: [25]; LSU: [4] |
| Daldinia concentrica | CBS 113277 | Germany | AY616683 | KY610434 | KY624243 | KC977274 | ITS: [26]; TUB2: [27]; [4] |
| Dematophora necatrix | CBS349.36 | Argentina | AY909001 | KF719204 | KY624275 | KY624310 | ITS: [5]; LSU: [4] |
| De. buxi | JDR99 | France | GU300070 | N/A | GQ844780 | GQ470228 | [23] |
| Euepixylon sphaeriostomum | JDR261 | USA | GU292821 | N/A | GQ844774 | GQ470224 | [23] |
| Graphostroma platystomum | CBS 270.87 | France | JX658535 | DQ836906 | KY624296 | HG934108 | ITS: [28]; LSU: [29]; RPB2: [30]; TUB2: [4] |
| Hypocopra anomala | TTI-000339 | in press | N/A | MT903245 | MT901033 | MT901030 | this study |
| Hypoc. dolichopoda | TTI-0310 | USA | N/A | MT903247 | MT901035 | N/A | this study |
| Hypoc. rostrata | TTI-000009 | USA | MT896134 | MT903246 | MT901034 | MT901031 | this study |
| Hypoxylon fragiforme | MUCL 51264/ET | Germany | KC477229 | KM186295 | KM186296 | KX271282 | ITS: [31]; RPB2: [3,4] |
| Kretzschmaria deusta | CBS163693 | Germany | KC477237 | KY610458 | KY624227 | KX271251 | [4] |
| Nemania abortiva | BISH 467/HT | USA | GU292816 | N/A | GQ844768 | GQ470219 | [23] |
| Nem. beaumontii | HAST405 | Martinique | GU292819 | N/A | GQ844772 | GQ470222 | [23] |
| Nem. bipapillata | HAST90080610 | Taiwan | GU292818 | N/A | GQ844771 | GQ470221 | [23] |
| Nem. maritima | DSM104968 | France | KY610414 | KY610414 | N/A | N/A | [4] |
| Nem. primolutea | HAST91102001/HT | Taiwan | EF026121 | N/A | GQ844767 | EF025607 | [23] |
| Podosordaria leporina | TTI-0312 | USA | N/A | MT903244 | MT901032 | MT901029 | this study |
| Podos. mexicana | WSP176 | Mexico | GU324762 | N/A | GQ853039 | GQ844840 | [23] |
| Podos. muli | WSP 167/HT | Mexico | GU324761 | N/A | GQ853038 | GQ844839 | [23] |
| Poronia pileiformis | WSP88113001/ET | Taiwan | GU324760 | N/A | GQ853037 | GQ502720 | [23] |
| Poronia punctata | CBS656.78 | Australia | KT281904 | KY610496 | KY624278 | KX271281 | ITS: [32]; [4] |
| Rosellinia aquila | MUCL51703 | France | KY610392 | KY610460 | KY624285 | KX271253 | [4] |
| Ros. corticium | MUCL51693 | France | KY610393 | KY610461 | KY624229 | KX271254 | [4] |
| Sarcoxylon compunctum | CBS359.61 | South Africa | KT281903 | KY610462 | KY624230 | KX271255 | ITS: [32]; [4] |
| Stilbohypoxylon elaeidicola | YMJ173 | French Guiana | EF026148 | N/A | GQ844826 | EF025616 | [23] |
| Stilboh. quisquiliarum | YMJ 172 | French Guiana | EF026119 | N/A | GQ853020 | EF025605 | [23] |
| Stromatoneurospora phoenix | BCC82040 | Thailand | MT703666 | MT735133 | MT742605 | MT700438 | this study |
| Stromatoneurospora phoenix | BCC82041 | Thailand | MT703667 | MT735134 | MT742606 | MT700439 | this study |
| Stromaton. phoenix | F-160, 834 | Mexico | AY909004 | N/A | N/A | N/A | [5] |
| Stromaton. phoenix | OTU_33 | China | MH430290 | N/A | N/A | N/A | [6] |
| Xylaria acuminatilongissima | HAST95060506/HT | Taiwan | EU178738 | EU178738 | EU178738 | EU178738 | [23] |
| Xyl. adscendens | JDR 865 | Thailand | GU322432 | N/A | GQ844818 | GQ487709 | [23] |
| Xyl. allantoidea | HAST 94042903 | Taiwan | GU324743 | N/A | GQ848356 | GQ502692 | [23] |
| Xyl. allantoidea | BCC22746 | Thailand | MT703671 | MT735141 | MT742610 | N/A | this study |
| Xyl. arbuscula | CBS126415 | Thailand | MH864101 | KY610463 | KY624287 | KX271257 | ITS: [33]; LSU, RPB2, TUB2: [4] |
| Xyl. bambusicola | BCC22739 | Thailand | MT710944 | MT735135 | MT742614 | N/A | this study |
| Xyl. bambusicola | WSP205/HT | Thailand | EF026123 | N/A | GQ844802 | AY951762 | [23] |
| Xyl. brunneovinosa | HAST720/HT | Taiwan | EU179862 | N/A | GQ853023 | GQ502706 | [23] |
| Xyl. cubensis | HAST 515 | Martinique | GU373810 | N/A | GQ848366 | GQ502701 | [23] |
| Xyl. cubensis | BCC20646 | Thailand | MT703672 | MT735142 | MT742611 | N/A | this study |
| Xyl discolor | HAST131023 | USA | JQ087405 | N/A | JQ087411 | JQ087414 | [34] |
| Xyl. grammica | BCC20655 | Thailand | MT703670 | MT735138 | MT742609 | N/A | this study |
| Xyl. grammica | isolate 479 | Taiwan | GU300097 | N/A | GQ844813 | GQ487704 | [23] |
| Xyl. hypoxylon | CBS122620/ET | Sweden | KY610407 | KY610495 | KY624231 | KX271279 | [23] |
| Xyl. ianthinovelutina | HAST 553 | Martinique | GU322441 | N/A | GQ844828 | GQ495934 | [23] |
| Xyl. multiplex | HAST 580 | Martinique | GU300098 | N/A | GQ844814 | GQ487705 | [23] |
| Xyl. polymorpha | MUCL49884 | France | KY610408 | KY610464 | KY624288 | KX271280 | [4] |
| Xyl. telfairii | BCC23019 | Thailand | MT703674 | MT735139 | MT742613 | N/A | this study |
| Xyl. telfairii | HAST 90081901 | Thailand | GU324738 | N/A | GQ848351 | GQ502687 | [23] |
| Pos 1 | 1 | 2 | ||
|---|---|---|---|---|
| δC, Mult 2 | δH, Mult 2 | δC, Mult 2 | δH, Mult 2 | |
| 1 | 33.5, CH2 | 2.37, m 2.19, m | 32.1, CH2 | 2.28, m 2.16, m |
| 2 | 29.7, CH2 | 1.99, m 1.41, m | 28.6, CH2 | 1.92, m 1.31, m |
| 3 | 31.1, CH2 | 1.62, m | 29.7, CH2 | 1.53, m |
| 4 | 41.1, CH | 1.86, m | 39.8, CH | 1.69, m |
| 5 | 42.4, C | 40.9, C | ||
| 6 | 70.3, CH | 3.43, s | 65.2, CH | 3.58, s |
| 7 | 62.8, C | - | 59.9, C | - |
| 8 | 192.6, C | - | 191.4, C | - |
| 9 | 120.6, CH | 5.69, s | 119.3, CH | 5.72, s |
| 10 | 169.0, C | - | 169.1, C | - |
| 11 | 146.7, C | - | 58.3, C | - |
| 12 | 63.7, CH2 | 4.26, d 4.17, d | 60.4, CH2 | 3.42, ddd (11.90, 4.88, 0.70) |
| 13 | 112.2, CH2 | 5.22, q (1.37) 5.13, dt (1.91, 1.03) | 46.4, CH2 | 2.65, dd (11.98, 4.81) 2.55, dd (5.49, 0.61) |
| 14 | 15.6, CH3 | 1.05, d (6.71) | 15.0, CH3 | 0.98, d (6.71) |
| 15 | 15.5, CH3 | 1.28, s | 14.9, CH3 | 1.18, s |
| 12-OH | - | - | - | 4.88, dd (7.32, 4.88) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becker, K.; Wongkanoun, S.; Wessel, A.-C.; Bills, G.F.; Stadler, M.; Luangsa-ard, J.J. Phylogenetic and Chemotaxonomic Studies Confirm the Affinities of Stromatoneurospora phoenix to the Coprophilous Xylariaceae. J. Fungi 2020, 6, 144. https://doi.org/10.3390/jof6030144
Becker K, Wongkanoun S, Wessel A-C, Bills GF, Stadler M, Luangsa-ard JJ. Phylogenetic and Chemotaxonomic Studies Confirm the Affinities of Stromatoneurospora phoenix to the Coprophilous Xylariaceae. Journal of Fungi. 2020; 6(3):144. https://doi.org/10.3390/jof6030144
Chicago/Turabian StyleBecker, Kevin, Sarunyou Wongkanoun, Anna-Charleen Wessel, Gerald F. Bills, Marc Stadler, and J. Jennifer Luangsa-ard. 2020. "Phylogenetic and Chemotaxonomic Studies Confirm the Affinities of Stromatoneurospora phoenix to the Coprophilous Xylariaceae" Journal of Fungi 6, no. 3: 144. https://doi.org/10.3390/jof6030144
APA StyleBecker, K., Wongkanoun, S., Wessel, A. -C., Bills, G. F., Stadler, M., & Luangsa-ard, J. J. (2020). Phylogenetic and Chemotaxonomic Studies Confirm the Affinities of Stromatoneurospora phoenix to the Coprophilous Xylariaceae. Journal of Fungi, 6(3), 144. https://doi.org/10.3390/jof6030144