Structural Characterization of the S-glycosylated Bacteriocin ASM1 from Lactobacillus plantarum
Abstract
:1. Introduction
2. Results
2.1. NMR Assignment and Post-Translational Modifications of ASM1
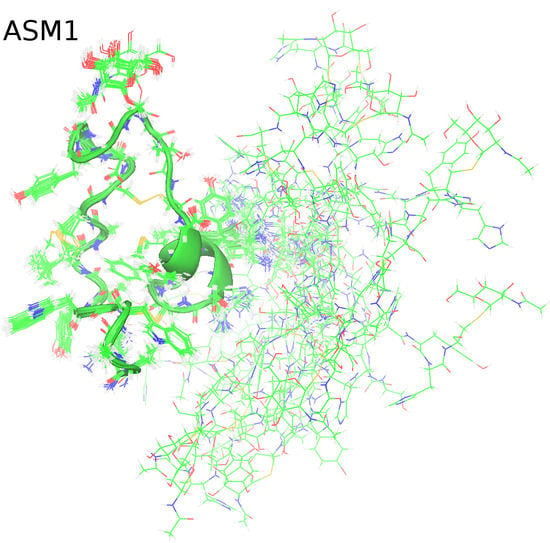
2.2. Solution NMR Structure of ASM1
2.3. Circular dichroism (CD) Spectroscopy
3. Discussion
4. Materials and Methods
4.1. NMR Spectroscopy
4.2. CD Spectroscopy
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Heng, N.C.K.; Tagg, J.R. What’s in a name? Class distinction for bacteriocins. Nat. Rev. Microbiol. 2016, 4, 160. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalban-Lopez, M.; Mu, D.; Kuipers, O.P. Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Norris, G.E.; Patchett, M.L. The glycocins: In a class of their own. Curr. Opin. Struct. Biol. 2016, 40, 112–119. [Google Scholar] [CrossRef]
- Venugopal, H.; Edwards, P.J.B.; Schwalbe, M.; Claridge, J.K.; Libich, D.S.; Stepper, J.; Loo, T.; Patchett, M.L.; Norris, G.E.; Pascal, S.M. Structural, dynamic, and chemical characterization of a novel S-glycosylated bacteriocin. Biochemistry 2011, 50, 2748–2755. [Google Scholar] [CrossRef]
- Garcia De Gonzalo, C.V.; Zhu, L.; Oman, T.J.; van der Donk, W.A. NMR structure of the S-linked glycopeptide sublancin 168. ACS Chem. Biol. 2014, 9, 796–801. [Google Scholar] [CrossRef]
- Stepper, J.; Shastri, S.; Loo, T.S.; Preston, J.C.; Novak, P.; Man, P.; Moore, C.H.; Havlicek, V.; Patchett., M.L.; Norris, G.E. Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett. 2011, 585, 645–650. [Google Scholar] [CrossRef] [Green Version]
- Hata, T.; Tanaka, R.; Ohmomo, S. Isolation and characterization of plantaricin ASM1: A new bacteriocin produced by Lactobacillus plantarum A-1. Int. J. Food Microbiol. 2010, 137, 94–99. [Google Scholar] [CrossRef]
- Main, P.; Hata, T.; Loo, T.S.; Man, P.; Novak, P.; Havlίček, V.; Norris, G.E.; Patchett, M.L. Bacteriocin ASM1 is an O/S-diglycosylated plasmid-encoded homologue of glycocin F. FEBS Lett. 2019. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.S.; Wilkinson, B.L.; O’Connell, M.R.; Mackay, J.P.; Matthews, J.M.; Payne, R.J. Synthesis of the bacteriocin glycopeptide sublancin 168 and S-glycosylated variants. Org. Lett. 2012, 14, 1910–1913. [Google Scholar] [CrossRef] [PubMed]
- Chagot, B.; Pimentel, C.; Dai, L.; Pil, J.; Tytgat, J.; Nakajima, T.; Corzo, G.; Darbon, H.; Ferrat, G. An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis. Biochem. J. 2005, 388, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Moller, C.; Rahmankhah, S.; Lauer-Fields, J.; Bubis, J.; Fields, G.B.; Mari, F. A novel conotoxin framework with a helix-loop-helix (Cs alpha/alpha) fold. Biochemistry 2005, 44, 15986–15996. [Google Scholar] [CrossRef] [PubMed]
- Nolde, S.B.; Vassilevski, A.A.; Rogozhin, E.A.; Barinov, N.A.; Balashova, T.A.; Samsonova, O.V.; Baranov, Y.V.; Feofanov, A.V.; Egorov, T.A.; Arseniev, A.S.; et al. Disulfide-stabilized helical hairpin structure and activity of a novel antifungal peptide EcAMP1 from seeds of barnyard grass (Echinochloa crus-galli). J. Biol. Chem. 2011, 286, 25145–25153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saucedo, A.L.; Flores-Solis, D.; Rodriguez de la Vega, R.C.; Ramirez-Cordero, B.; Hernandez-Lopez, R.; Cano-Sanchez, P.; Noriega Navarro, R.; Garcia-Valdes, J.; Coronas-Valderrama, F.; de Roodt, A.; et al. New tricks of an old pattern: Structural versatility of scorpion toxins with common cysteine spacing. J. Biol. Chem. 2012, 287, 12321–12330. [Google Scholar] [CrossRef] [Green Version]
- Güntert, P. Automatic NMR Structure Calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Defining solution conformations of small linear peptides. Annu. Rev. Biophys. Biophys. Chem. 1991, 60, 795–825. [Google Scholar] [CrossRef]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar] [CrossRef]
- Provencher, S.W.; Glockner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 1981, 20, 33–37. [Google Scholar] [CrossRef]
- Domard, A. Circular dichroism study on N-acetylglucosamineoligomers. Int. J. Biol. Macromol. 1986, 8, 243–246. [Google Scholar] [CrossRef]
- Alexander, P.A.; He, Y.; Chen, Y.; Orban, J.; Brayn, P.N. A minimal sequence code for switching protein structure and function. Proc. Natl. Acad. Sci. USA 2009, 106, 21149–21154. [Google Scholar] [CrossRef] [Green Version]
- Goroncy, A.K.; Koshiba, S.; Tochio, N.; Tomizawa, T.; Sato, M.; Inoue, M.; Watanabe, S.; Hayashizaki, Y.; Tanaka, A.; Kigawa, T.; et al. NMR solution structures of actin depolymerizing factor homology domains. Protein Sci. 2009, 18, 2384–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Iwahara, J.; Koshiba, S.; Tomizawa, T.; Tochio, N.; Güntert, P.; Kigawa, T.; Yokoyama, S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to the high-throughput NMR structure studies. J. Biomol. NMR 2007, 39, 31–52. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.; Blevins, R. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef]
- Kleywegt, G.J. Crystallographic refinement of ligand complexes. Acta Crystallogr. Sect. D 2007, 63, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Karpral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D 2010, 66, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Cornilescu, G.; Delaglio, F.; Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 1999, 13, 289–302. [Google Scholar] [CrossRef]
- Cordier, F.; Nisius, L.; Dingley, A.J.; Grzesiek, S. Direct detection of N−H O=C hydrogen bonds in biomolecules by NMR spectroscopy. Nat. Protoc. 2008, 3, 235–241. [Google Scholar] [CrossRef]
- Blackledge, M. NMR provides evidence for dynamic hydrogen bonding in proteins. Protein Sci. 2007, 16, 1247–1248. [Google Scholar] [CrossRef]
- Singh, S. Excess Gibbs for binary mixtures of acetonitrile with acetic acid, propionic acid, isobutyric acid, and trimethylacetic acid. Can. J. Chem. 1991, 69, 2117–2121. [Google Scholar] [CrossRef] [Green Version]
- Pavlov, S.Y.; Pavlova, S.P.; Kirnos, A.B. Azeotropes of acetonitrile with C5 hydrocarbons and with water. Zhurnal Pikladnoi Khimii 1966, 39, 1555–1559. [Google Scholar]
- Gekk, K.; Ohmae, E.; Kameyama, K.; Takagi, T. Acetonitrile-protein interactions: Amino acid solubility and preferential solvation. Biochim. Biophys. Acta 1998, 1387, 195–205. [Google Scholar] [CrossRef]
- Güntert, P. Automated NMR structure calculation. Prog. Nucl. Magn. Reson. Spectrosc. 2003, 43, 105–125. [Google Scholar] [CrossRef]
- Güntert, P.; Mumenthaler, C.; Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program CYANA. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Rullmann, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMol Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2008. [Google Scholar]
- Koradi, R.; Billeter, M.; Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Available online: http://www.pdb (accessed on 2 March 2020).
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Lobley, A.; Whitmore, L.; Wallace, B.A. DICHROWEB: An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 2002, 18, 211–212. [Google Scholar] [CrossRef] [Green Version]








| GlcNAc (Attached to Ser-18) | GlcNAc (Attached to Cys-43) | |||
|---|---|---|---|---|
| Constituent | δ(1H)/ppm | δ(13C)/ppm | δ(1H)/ppm | δ(13C)/ppm |
| 1 | 4.479 | 100.952 | 4.508 | 83.723 |
| 2 | 3.653 | 55.404 | 3.691 | 54.452 |
| 3 | 3.485 | 73.788 | 3.447 | 75.184 |
| 4 | 3.345 | 69.747 | 3.350 | 69.939 |
| 5 | 3.388 | 75.898 | 3.352 | 80.108 |
| 6 | 3.627 | 61.050 | 3.808 | 61.226 |
| 6′ | 3.627 | 3.808 | ||
| Me | 1.994 | 22.413 | 1.900 | 22.134 |
| Input Data | Without H-Bond Restraints | With H-Bond Restraints |
|---|---|---|
| Completeness of NMR assignments (%) | ||
| - of backbone amide protons and aliphatic protons | 99.6% | 99.6% |
| Total assigned atoms | 438 | 438 |
| Dihedral angle constraints (ф, ψ) | 33 (17,16) | 33 (17,16) |
| Total number of 1H-1H-NOESY derived peaks | 1768 | 1768 |
| CYANA-version used | 3.0 | 3.0 |
| Restraints for H-bonds α-helices 5–12, 21–28 | 0 | 16 |
| Output Data | ||
| CYANA target function value (Å2) | 0.68 | 12.00 |
| NOE-derived distance constraints | 577 | 569 |
| - Intraresidual (|i − j| < 1) | 323 | 295 |
| - Medium-range (1 < i − j| < 5) | 138 | 168 |
| - Long-range (|i − j| > 5) | 116 | 106 |
| Unassigned NOE peaks | 72 | 98 |
| RMS deviation from idealized geometry | ||
| - Bond lengths (Å) | 0.004 | 0.005 |
| - Bond angles (degrees) | 0.7 | 0.8 |
| RMS deviation from averaged coordinates (Å) a | ||
| - Backbone | 0.17 | 0.11 |
| - Heavy atoms | 0.38 | 0.42 |
| Ramachandran analysis (%) a | ||
| - Most favoured | 52.8 | 48.4 |
| - Additionally allowed | 38.8 | 40.5 |
| - Generously allowed | 8.4 | 11.1 |
| - Disallowed | 0.0 | 0.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goroncy, A.K.; Loo, T.S.; Koolaard, A.M.; Patchett, M.L.; Norris, G.E. Structural Characterization of the S-glycosylated Bacteriocin ASM1 from Lactobacillus plantarum. Magnetochemistry 2020, 6, 16. https://doi.org/10.3390/magnetochemistry6010016
Goroncy AK, Loo TS, Koolaard AM, Patchett ML, Norris GE. Structural Characterization of the S-glycosylated Bacteriocin ASM1 from Lactobacillus plantarum. Magnetochemistry. 2020; 6(1):16. https://doi.org/10.3390/magnetochemistry6010016
Chicago/Turabian StyleGoroncy, Alexander K., Trevor S. Loo, Adrian M. Koolaard, Mark L. Patchett, and Gillian E. Norris. 2020. "Structural Characterization of the S-glycosylated Bacteriocin ASM1 from Lactobacillus plantarum" Magnetochemistry 6, no. 1: 16. https://doi.org/10.3390/magnetochemistry6010016
APA StyleGoroncy, A. K., Loo, T. S., Koolaard, A. M., Patchett, M. L., & Norris, G. E. (2020). Structural Characterization of the S-glycosylated Bacteriocin ASM1 from Lactobacillus plantarum. Magnetochemistry, 6(1), 16. https://doi.org/10.3390/magnetochemistry6010016