Reducing Inert Materials for Optimal Cell–Cell and Cell–Matrix Interactions within Microphysiological Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microfluidic Device Fabrication and Validation
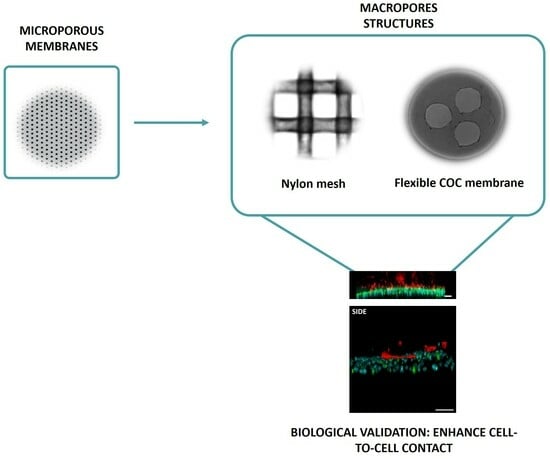
2.1.1. Mesh Device
2.1.2. Macropore Device
2.2. Cell Culture
2.3. Spheroid Generation
2.4. Collagen Hydrogel Preparation
2.5. Microfluidic Device Seeding
2.5.1. Epithelium Generation
2.5.2. Migration Assays with U-87 Spheroids
2.5.3. Co-Culture Assays with Epithelial and Fibroblasts Cells
2.6. Membrane Permeability Evaluation
2.7. Immunostaining
2.8. Image Analysis
2.9. Graph Analysis
2.10. Statistical Analysis
3. Results
3.1. Manufacturing of the Microfluidic Devices
3.2. Validation of the Microfluidic Devices
3.2.1. Epithelium Generation
3.2.2. Migration Assays
3.2.3. Co-Culture Assays with Epithelial Cells and Fibroblasts
3.3. Permeability Assessment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, J.X.J.; Hoshino, K. Microfluidics and Micro Total Analytical Systems. Mol. Sensors Nanodevices 2014, 103–168. [Google Scholar] [CrossRef]
- Convery, N.; Gadegaard, N. 30 Years of Microfluidics. Micro Nano Eng. 2019, 2, 76–91. [Google Scholar] [CrossRef]
- Wang, K.; Man, K.; Liu, J.; Liu, Y.; Chen, Q.; Zhou, Y.; Yang, Y. Microphysiological Systems: Design, Fabrication, and Applications. ACS Biomater. Sci. Eng. 2020, 6, 3231–3257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, G.; Wu, J.; Liu, X.; Fan, Y.; Chen, J.; Wallace, G.; Gu, Q. Microphysiological Constructs and Systems: Biofabrication Tactics, Biomimetic Evaluation Approaches, and Biomedical Applications. Small Methods 2023, 8, 2300685. [Google Scholar] [CrossRef]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022, 23, 467–491. [Google Scholar] [CrossRef] [PubMed]
- Pasqualini, F.S.; Emmert, M.Y.; Parker, K.K.; Hoerstrup, S.P. Organ Chips: Quality Assurance Systems in Regenerative Medicine. Clin. Pharmacol. Ther. 2017, 101, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.M.; de Haan, P.; Ronaldson-Bouchard, K.; Kim, G.-A.; Ko, J.; Rho, H.S.; Chen, Z.; Habibovic, P.; Jeon, N.L.; Takayama, S.; et al. A guide to the organ-on-a-chip. Nat. Rev. Methods Prim. 2022, 2, 1–29. [Google Scholar] [CrossRef]
- Joseph, X.; Akhil, V.; Arathi, A.; Mohanan, P.V. Comprehensive Development in Organ-On-A-Chip Technology. J. Pharm. Sci. 2022, 111, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Monteduro, A.G.; Rizzato, S.; Caragnano, G.; Trapani, A.; Giannelli, G.; Maruccio, G. Organs-on-chips technologies—A guide from disease models to opportunities for drug development. Biosens. Bioelectron. 2023, 231, 115271. [Google Scholar] [CrossRef]
- Pasman, T.; Grijpma, D.; Stamatialis, D.; Poot, A. Flat and microstructured polymeric membranes in organs-on-chips. J. R. Soc. Interface 2018, 15, 20180351. [Google Scholar] [CrossRef]
- Salminen, A.T.; Zhang, J.; Madejski, G.R.; Khire, T.S.; Waugh, R.E.; McGrath, J.L.; Gaborski, T.R. Ultrathin Dual-Scale Nano- and Microporous Membranes for Vascular Transmigration Models. Small 2019, 15, e1804111. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Sun, M.; Zhang, J.; Fu, W.; Hu, R.; Liu, D.; Liu, W. Large-scale investigation of single cell activities and response dynamics in a microarray chip with a microfluidics-fabricated microporous membrane. Analyst 2021, 146, 4303–4313. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.H.; Mireles, M.; Kwarta, B.J.; Gaborski, T.R. Use of porous membranes in tissue barrier and co-culture models. Lab Chip 2018, 18, 1671–1689. [Google Scholar] [CrossRef]
- Moeendarbary, E.; Harris, A.R. Cell mechanics: Principles, practices, and prospects. Wiley Interdiscip. Rev. Syst. Biol. Med. 2014, 6, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Hastings, J.F.; Skhinas, J.N.; Fey, D.; Croucher, D.R.; Cox, T.R. The extracellular matrix as a key regulator of intracellular signalling networks. Br. J. Pharmacol. 2019, 176, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K.; Dandara, C. The Extracellular Matrix: Its Composition, Function, Remodeling, and Role in Tumorigenesis. Biomimetics 2023, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Berrier, A.L.; Yamada, K.M. Cell–Matrix Adhesion. Cell. Physiol. 2007, 213, 565–573. [Google Scholar] [CrossRef]
- Yamada, K.M.; Doyle, A.D.; Lu, J. Cell–3D matrix interactions: Recent advances and opportunities. Trends Cell Biol. 2022, 32, 883–895. [Google Scholar] [CrossRef]
- Watt, F.M.; Fujiwara, H. Cell-extracellular matrix interactions in normal and diseased skin. Cold Spring Harb Perspect Biol. 2011, 3, a005124. [Google Scholar] [CrossRef]
- Nishida-Aoki, N.; Gujral, T.S. Review Emerging approaches to study cell–cell interactions in tumor microenvironment. Oncotarget 2019, 10, 785–797. [Google Scholar] [CrossRef]
- Muschler, J.; Streuli, C.H. Cell-matrix interactions in mammary gland development and breast cancer. Cold Spring Harb. Perspect. Biol. 2010, 2, a003202. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.J.; Jones, M.C. ScienceDirect Cell cycle control by cell-matrix interactions. Curr. Opin. Cell Biol. 2024, 86, 102288. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Banerjee, S.; Baker, G.W.; Kuo, C.Y.; Chowdhury, I. The Mammary Gland: Basic Structure and Molecular Signaling during Development. Int. J. Mol. Sci. 2022, 23, 3883. [Google Scholar] [CrossRef] [PubMed]
- Vela-Alcántara, A.; Tamariz, E. Cell-Cell and Cell-Matrix Interactions during Axons Guidance. In Neurons—Dendrites and Axons; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- it4ip S.A. 1, Avenue Jean-Etienne Lenoir 1348 Louvain-la-Neuve (Belgium). 2023, Volume 32, p. 226114. Available online: https://www.it4ip.be/ (accessed on 19 February 2024).
- Castro-Abril, H.; Heras, J.; del Barrio, J.; Paz, L.; Alcaine, C.; Aliácar, M.P.; Garzón-Alvarado, D.; Doblaré, M.; Ochoa, I. The Role of Mechanical Properties and Structure of Type I Collagen Hydrogels on Colorectal Cancer Cell Migration. Macromol. Biosci. 2023, 23, e202300108. [Google Scholar] [CrossRef] [PubMed]
- Ayuso, J.M.; Virumbrales-Munoz, M.; McMinn, P.H.; Rehman, S.; Gomez, I.; Karim, M.R.; Trusttchel, R.; Wisinski, K.B.; Beebe, D.J.; Skala, M.C. Tumor-on-A-chip: A microfluidic model to study cell response to environmental gradients. Lab Chip 2019, 19, 3461–3471. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Carro, E.; Salomon-Cambero, R.; Armero, L.; Castro-Abril, H.A.; Ayensa-Jiménez, J.; Martínez, M.A.; Ochoa, I.; Alcaine, C.; García, I.; Ciriza, J. Nanoparticles Stokes radius assessment through permeability coefficient determination within a new stratified epithelium on-chip model. Artif. Cells Nanomed. Biotechnol. 2023, 51, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Olaizola-Rodrigo, C.; Palma-Florez, S.; Ranđelović, T.; Bayona, C.; Ashrafi, M.; Samitier, J.; Lagunas, A.; Mir, M.; Doblaré, M.; Ochoa, I.; et al. Tuneable hydrogel patterns in pillarless microfluidic devices. Lab Chip 2024, 24, 2094–2106. [Google Scholar] [CrossRef] [PubMed]
- Pires, C.L.; Praça, C.; Martins, P.A.T.; de Carvalho, A.L.M.B.; Ferreira, L.; Marques, M.P.M.; Moreno, M.J. Re-use of caco-2 monolayers in permeability assays—Validation regarding cell monolayer integrity. Pharmaceutics 2021, 13, 1563. [Google Scholar] [CrossRef] [PubMed]
- Van Poll, M.L.; Zhou, F.; Ramstedt, M.; Hu, L.; Huck, W.T.S. A self-assembly approach to chemical micropatterning of poly(dimethylsiloxane). Angew. Chem. Int. Ed. 2007, 46, 6634–6637. [Google Scholar] [CrossRef]
- Berthier, E.; Young, E.W.K.; Beebe, D. Engineers are from PDMS-land, biologists are from polystyrenia. Lab Chip 2012, 12, 1224–1237. [Google Scholar] [CrossRef]
- Kuddannaya, S.; Bao, J.; Zhang, Y. Enhanced in Vitro Biocompatibility of Chemically Modified Poly(dimethylsiloxane) Surfaces for Stable Adhesion and Long-term Investigation of Brain Cerebral Cortex Cells. ACS Appl. Mater. Interfaces 2015, 7, 25529–25538. [Google Scholar] [CrossRef] [PubMed]
- Samarth, N.; Kamble, V.; Rane, A.V.; Abitha, V.K.; Gohatre, O.; Kanny, K. Mechanical and Dynamic Mechanical Properties of Polyurethane Blends and Interpenetrating Polymer Networks; Elsevier Inc.: Amsterdam, The Netherlands, 2017. [Google Scholar]
- McMillan, A.H.; Thomée, E.K.; Dellaquila, A.; Nassman, H.; Segura, T.; Lesher-Pérez, S.C. Rapid fabrication of membrane-integrated thermoplastic elastomer microfluidic devices. Micromachines 2020, 11, 731. [Google Scholar] [CrossRef] [PubMed]
- Eain, M.M.G.; Baginska, J.; Greenhalgh, K.; Fritz, J.V.; Zenhausern, F.; Wilmes, P. Engineering Solutions for Representative Models of the Gastrointestinal Human-Microbe Interface. Engineering 2017, 3, 60–65. [Google Scholar] [CrossRef]
- van der Helm, M.W.; Odijk, M.; Frimat, J.-P.; van der Meer, A.D.; Eijkel, J.C.; Berg, A.v.D.; Segerink, L.I. Direct quantification of transendothelial electrical resistance in organs-on-chips. Biosens. Bioelectron. 2016, 85, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.I.; Abaci, H.E.; Shuler, M.L. Microfluidic blood–brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 2017, 114, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Maschmeyer, I.; Hasenberg, T.; Jaenicke, A.; Lindner, M.; Lorenz, A.K.; Zech, J.; Garbe, L.-A.; Sonntag, F.; Hayden, P.; Ayehunie, S.; et al. Chip-based human liver-intestine and liver-skin co-cultures—A first step toward systemic repeated dose substance testing in vitro. Eur. J. Pharm. Biopharm. 2015, 95, 77–87. [Google Scholar] [CrossRef]
- Krishnan, S.G.P.; Kulkarni, S.T. Polyester resins. Polyest. Polyam. 2008, 3–40. [Google Scholar] [CrossRef]
- East, A.J.; Golder, M.; Makhija, S. Polyesters, Thermoplastic. Kirk-Othmer Encycl. Chem. Technol. 2000, 7, 504–529. [Google Scholar] [CrossRef]
- Moradi, E.; Jalili-Firoozinezhad, S.; Solati-Hashjin, M. Microfluidic organ-on-a-chip models of human liver tissue. Acta Biomater. 2020, 116, 67–83. [Google Scholar] [CrossRef]
- Polidoro, M.A.; Ferrari, E.; Marzorati, S.; Lleo, A.; Rasponi, M. Experimental liver models: From cell culture techniques to microfluidic organs-on-chip. Liver Int. 2021, 41, 1744–1761. [Google Scholar] [CrossRef]
- Persson, H.; Park, S.; Mohan, M.; Cheung, K.K.; Simmons, C.A.; Young, E.W.K. Rapid assembly of PMMA microfluidic devices with PETE membranes for studying the endothelium. Sensors Actuators B Chem. 2021, 356, 131342. [Google Scholar] [CrossRef]
- Cipitria, A.; Skelton, A.; Dargaville, T.R.; Dalton, P.D.; Hutmacher, D.W. Design, fabrication and characterization of PCL electrospun scaffolds—A review. J. Mater. Chem. 2011, 21, 9419–9453. [Google Scholar] [CrossRef]
- Ongaro, A.E.; Keraite, I.; Liga, A.; Conoscenti, G.; Coles, S.; Schulze, H.; Bachmann, T.T.; Parvez, K.; Casiraghi, C.; Howarth, N.M.; et al. Laser Ablation of Poly(lactic acid) Sheets for the Rapid Prototyping of Sustainable, Single-Use, Disposable Medical Microcomponents. ACS Sustain. Chem. Eng. 2018, 6, 4899–4908. [Google Scholar] [CrossRef]
- Ongaro, A.E.; Di Giuseppe, D.; Kermanizadeh, A.; Crespo, A.M.; Mencattini, A.; Ghibelli, L.; Mancini, V.; Wlodarczyk, K.L.; Hand, D.P.; Martinelli, E.; et al. Polylactic is a Sustainable, Low Absorption, Low Autofluorescence Alternative to Other Plastics for Microfluidic and Organ-on-Chip Applications. Anal. Chem. 2020, 92, 6693–6701. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, H.; Nicolas, A.; Schavemaker, F.; Heijmans, J.P.M.; Bulst, M.; Trietsch, S.J.; Broek, L.J.v.D. Vascular inflammation on a chip: A scalable platform for trans-endothelial electrical resistance and immune cell migration. Front. Immunol. 2023, 14, 1118624. [Google Scholar] [CrossRef] [PubMed]
- Gjorevski, N.; Nikolaev, M.; Brown, T.E.; Mitrofanova, O.; Brandenberg, N.; DelRio, F.W.; Yavitt, F.M.; Liberali, P.; Anseth, K.S.; Lutolf, M.P. Tissue geometry drives deterministic organoid patterning. Science 2022, 375, eaaw9021. [Google Scholar] [CrossRef]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknaes, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef] [PubMed]
- Fomby, P.; Cherlin, A.J.; Hadjizadeh, A.; Doillon, C.J.; Sueblinvong, V.; Weiss, D.J.; Bates, J.H.T.; Gilbert, T.; Liles, W.C.; Lutzko, C.; et al. Stem cells and cell therapies in lung biology and diseases: Conference report. Ann. Am. Thorac. Soc. 2010, 12, 181–204. [Google Scholar]
- Metz, C.; Döger, R.; Riquelme, E.; Cortés, P.; Holmes, C.; Shaughnessy, R.; Oyanadel, C.; Grabowski, C.; González, A.; Soza, A. Galectin-8 promotes migration and proliferation and prevents apoptosis in U87 glioblastoma cells. Biol. Res. 2016, 49, 1–10. [Google Scholar] [CrossRef]
- Yang, J.-T.; Lee, I.-N.; Lu, F.-J.; Chung, C.-Y.; Lee, M.-H.; Cheng, Y.-C.; Chen, K.-T.; Chen, C.-H. Propyl Gallate Exerts an Antimigration Effect on Temozolomide-Treated Malignant Glioma Cells through Inhibition of ROS and the NF-κB Pathway. J. Immunol. Res. 2017, 2017, 1–12. [Google Scholar] [CrossRef]
- Huang, W.; Ding, X.; Ye, H.; Wang, J.; Shao, J.; Huang, T. Hypoxia enhances the migration and invasion of human glioblastoma U87 cells through PI3K/Akt/mTOR/HIF-1α pathway. Neuroreport 2018, 29, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- van Duinen, V.; Trietsch, S.J.; Joore, J.; Vulto, P.; Hankemeier, T. Microfluidic 3D cell culture: From tools to tissue models. Curr. Opin. Biotechnol. 2015, 35, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Sympson, C.J.; Talhouk, R.S.; Alexander, C.M.; Chin, J.R.; Clift, S.M.; Bissell, M.J.; Werb, Z. Targeted expression of stromelysin-1 in mammary gland provides evidence for a role of proteinases in branching morphogenesis and the requirement for an intact basement membrane for tissue-specific gene expression. J. Cell Biol. 1994, 125, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, R. In Vitro Methods for Measuring the Permeability of Cell Monolayers. Methods Protoc. 2022, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, A.P.; Han, H.C. Biomechanics of Cardiac Function. Compr Physiol. 2015, 5, 1623–1644. [Google Scholar] [CrossRef] [PubMed]
- Eskandari, M.; Nordgren, T.M.; O’Connell, G.D. Mechanics of pulmonary airways: Linking structure to function through constitutive modeling, biochemistry, and histology. Acta Biomater. 2019, 97, 513–523. [Google Scholar] [CrossRef]
- Divertie, M.B.; Cassan, S.M.; Brown, A.L. Ultrastructural morphometry of the blood air barrier in pulmonary sarcoidosis. Chest 1976, 69, 154–157. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olaizola-Rodrigo, C.; Castro-Abril, H.; Perisé-Badía, I.; Pancorbo, L.; Ochoa, I.; Monge, R.; Oliván, S. Reducing Inert Materials for Optimal Cell–Cell and Cell–Matrix Interactions within Microphysiological Systems. Biomimetics 2024, 9, 262. https://doi.org/10.3390/biomimetics9050262
Olaizola-Rodrigo C, Castro-Abril H, Perisé-Badía I, Pancorbo L, Ochoa I, Monge R, Oliván S. Reducing Inert Materials for Optimal Cell–Cell and Cell–Matrix Interactions within Microphysiological Systems. Biomimetics. 2024; 9(5):262. https://doi.org/10.3390/biomimetics9050262
Chicago/Turabian StyleOlaizola-Rodrigo, Claudia, Héctor Castro-Abril, Ismael Perisé-Badía, Lara Pancorbo, Ignacio Ochoa, Rosa Monge, and Sara Oliván. 2024. "Reducing Inert Materials for Optimal Cell–Cell and Cell–Matrix Interactions within Microphysiological Systems" Biomimetics 9, no. 5: 262. https://doi.org/10.3390/biomimetics9050262
APA StyleOlaizola-Rodrigo, C., Castro-Abril, H., Perisé-Badía, I., Pancorbo, L., Ochoa, I., Monge, R., & Oliván, S. (2024). Reducing Inert Materials for Optimal Cell–Cell and Cell–Matrix Interactions within Microphysiological Systems. Biomimetics, 9(5), 262. https://doi.org/10.3390/biomimetics9050262