Inadequate Sampling Frequency and Imprecise Taxonomic Identification Mask Results in Studies of Migratory Freshwater Fish Ichthyoplankton
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Methods
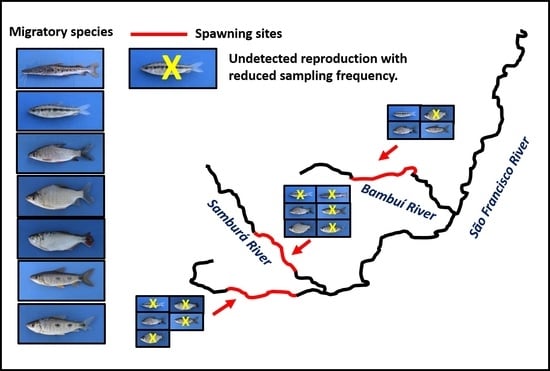
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Fish Species | ASV % | Migratory (Y/N) | Parental Care (Y/N) |
|---|---|---|---|
| ORDER CLUPEIFORMES | |||
| Family Engraulidae | |||
| Anchoviella vaillanti (Steindachner, 1908) | 0.06 | N | N |
| ORDER CHARACIFORMES | |||
| Family Crenuchidae | |||
| Characidium fasciatum Reinhardt, 1867 | <0.01 | N | N |
| Characidium zebra Eigenmann, 1909 | 0.07 | N | N |
| Family Parodontidae | |||
| Parodon hilarii Reinhardt, 1867 | <0.01 | N | N |
| Family Serrasalmidae | |||
| Myleus micans (Lütken, 1875) | 0.01 | N | N |
| Myleus sp. | <0.01 | N | N |
| Myloplus sp. | <0.01 | N | N |
| Serrasalmus sp. | <0.01 | N | Y |
| Family Anostomidae | |||
| Leporellus vittatus (Valenciennes, 1850) | 0.86 | N | N |
| Leporinus piau Fowler, 1941 | 0.04 | N | N |
| Leporinus taeniatus Lütken, 1875 | 2.26 | Y | N |
| Megaleporinus obtusidens (Valenciennes, 1837) | 0.90 | Y | N |
| Megaleporinus reinhardti (Lütken, 1875) | <0.01 | Y | N |
| Schizodon knerii (Steindachner, 1875) | 1.94 | N | N |
| Family Curimatidae | |||
| Curimatidae sp. | <0.01 | N | N |
| Family Prochilodontidae | |||
| Prochilodus argenteus Spix and Agassiz, 1829+ | 2.92 | Y | N |
| Prochilodus costatus Valenciennes, 1850 + | 2.88 | Y | N |
| Family Bryconidae | |||
| Brycon sp. | <0.01 | ? | N |
| Family Characidae | |||
| Astyanax scabripinnis (Jenyns, 1842) | 0.02 | N | N |
| Bryconamericus sp. | 0.26 | N | N |
| Piabina sp. | 0.11 | N | N |
| Psalidodon fasciatus (Cuvier, 1819) | 0.08 | N | N |
| Tetragonopterus chalceus Spix and Agassiz, 1829 | <0.01 | N | N |
| ORDER SILURIFORMES | |||
| Family Cetopsidae | |||
| Cetopsidae sp. | 0.05 | N | N |
| Family Doradidae | |||
| Doradidae sp. | <0.01 | N | N |
| Family Heptapteridae | |||
| Cetopsorhamdia iheringi Schubart and Gomes, 1959 | 0.57 | N | N |
| Imparfinis sp. | <0.01 | N | N |
| Family Pimelodidae | |||
| Bergiaria westermanni (Lütken, 1874) | 4.84 | N | N |
| Pimelodus fur (Lütken, 1874) | 1.29 | N | N |
| Pimelodus maculatus Lacepède, 1803 | 11.54 | N | N |
| Pimelodus pohli Ribeiro and Lucena, 2006 | 63.54 | N | N |
| Pseudoplatystoma corruscans (Spix and Agassiz, 1829) | 0.01 | Y | N |
| ORDER GYMNOTIFORMES | |||
| Family Sternopygidae | |||
| Eigenmannia sp. | <0.01 | N | N |
| ORDER PERCIFORMES | |||
| Family Sciaenidae | |||
| Pachyurus sp. | 3.89 | N | N |
| Plagioscion squamosissimus (Heckel, 1840) | <0.01 | N | N |
References
- Agostinho, A.A.; Pelicice, F.M.; Gomes, L.C. Dams and the fish fauna of the Neotropical region: Impacts and management related to diversity and fisheries. Braz. J. Biol. 2008, 68, 1119–1132. [Google Scholar] [CrossRef] [PubMed]
- Agostinho, A.A.; Thomaz, S.M.; Gomes, L.C. Conservação da biodiversidade em águas continentais do Brasil. Megadiversidade 2005, 1, 70–78. [Google Scholar]
- Godinho, A.L.; Pompeu, P.D.S. A importância dos ribeirões para os peixes de piracema. In Águas, Peixes e Pescadores do São Francisco da Minas Gerais; PUC Minas: Belo Horizonte, Brazil, 2003; pp. 361–372. [Google Scholar]
- Lopes, J.D.M.; Pompeu, P.S.; Alves, C.B.M.; Peressin, A.; Prado, I.G.; Suzuki, F.M.; Facchin, S.; Kalapothakis, E. The critical importance of an undammed river segment to the reproductive cycle of a migratory Neotropical fish. Ecol. Freshw. Fish 2019, 28, 302–316. [Google Scholar] [CrossRef]
- Pelicice, F.M.; Pompeu, P.S.; Agostinho, A.A. Large reservoirs as ecological barriers to downstream movements of Neotropical migratory fish. Fish Fishes 2015, 16, 697–715. [Google Scholar] [CrossRef]
- Nakatani, K.; Agostinho, A.A.; Baumgartner, G.; Bialetzki, A.; Sanches, P.V.; Makrakis, M.C.; Pavanelli, C.S. Ovos e Larvas de Peixes de Água Doce: Desenvolvimento e Manual de Identificação; EDUEM: Maringá, Brazil, 2001. [Google Scholar]
- Brasil, Ministério de Minas e Energia, Empresa de Pesquisa Energética. Plano Nacional de Energia 2050/Ministério de Minas e Energia; Empresa de Pesquisa Energética, MME/EPE: Brasília, Brazil, 2020. [Google Scholar]
- Pompeu, P.S.; Agostinho, A.A.; Pelicice, F.M. Existing and future challenges: The concept of successful fish passage in South America. River Res. Appl. 2012, 28, 504–512. [Google Scholar] [CrossRef]
- Orsi, M.L.; Almeida, F.S.; Swarça, A.C.; Claro-García, A.; Vianna, N.C.; Garcia, D.A.Z.; Bialetzki, A. Ovos, Larvas e Juvenis dos Peixes da Bacia do Rio Paranapanema uma Avaliação para a Conservação; Triunfal Gráfica e Editora: Assis, Brazil, 2016; 136p. [Google Scholar]
- Reynalte-Tataje, D.A.; Nakatani, K.; Fernandes, R.; Agostinho, A.A.; Bialetzki, A. Temporal distribution of ichthyoplankton in the Ivinhema River (Mato Grosso do Sul State/Brazil): Influence of environmental variables. Neotrop. Ichthyol. 2011, 9, 427–436. [Google Scholar] [CrossRef]
- Suzuki, F.M.; Pompeu, P.S. Influence of abiotic factors on ichthyoplankton occurrence in stretches with and without dams in the upper Grande River basin, south-eastern Brazil. Fish. Manag. Ecol. 2016, 23, 99–108. [Google Scholar] [CrossRef]
- Baumgartner, G.; Nakatani, K.; Gomes, L.C.; Bialetzki, A.; Sanches, P.V.; Makrakis, M.C. Fish larvae from the upper Paraná River: Do abiotic factors affect larval density? Neotrop. Ichthyol. 2008, 6, 551–558. [Google Scholar] [CrossRef]
- Reis, R.E.; Albert, J.S.; Di Dario, F.; Mincarone, M.M.; Petry, P.; Rocha, L.A. Fish biodiversity and conservation in South America. J. Fish Biol. 2016, 89, 12–47. [Google Scholar] [CrossRef]
- Nascimento, F.L.; Lima, C.A.R.M.A. Descrição das Larvas das Principais Espécies de Peixes Utilizadas pela Pesca, no Pantanal; Embrapa: Brasília, Brazil, 2000; 25p. [Google Scholar]
- Rizzo, E.; Sato, Y.; Barreto, B.P.; Godinho, H.P. Adhesiveness and surface patterns of eggs in neotropical freshwater teleosts. J. Fish Biol. 2002, 61, 615–632. [Google Scholar] [CrossRef]
- Pompeu, P.S.; Nogueira, L.B.; Godinho, H.P.; Martinez, C.B. Downstream passage of fish larvae and eggs through a small-sized reservoir, Mucuri River, Brazil. Zoology 2011, 28, 739–746. [Google Scholar] [CrossRef]
- Reynalte-Tataje, D.A.; Agostinho, A.A.; Bialetzki, A.; Hermes-Silva, S.; Fernandes, R.; Zaniboni-Filho, E. Spatial and temporal variation of the ichthyoplankton in a subtropical river in Brazil. Environ. Biol. Fishes 2012, 94, 403–419. [Google Scholar] [CrossRef]
- Becker, R.A.; Sales, N.G.; Santos, G.M.; Santos, G.B.; Carvalho, D.C. DNA barcoding and morphological identification of neotropical ichthyoplankton from the Upper Paraná and São Francisco. J. Fish Biol. 2015, 87, 159–168. [Google Scholar] [CrossRef]
- Leese, F.; Bouchez, A.; Abarenkov, K.; Altermatt, F.; Borja, Á.; Bruce, K.; Ekrem, T.; Čiampor, F., Jr.; Čiamporová-Zaťovičová, Z.; Costa, F.O.; et al. Why we need sustainable networks bridging countries, disciplines, cultures and generations for aquatic biomonitoring 2.0: A perspective derived from the DNAqua-Net COST action. Adv. Ecol. Res. 2018, 58, 63–99. [Google Scholar]
- Baird, D.J.; Hajibabaei, M. Biomonitoring 2.0: A new paradigm in ecosystem assessment made possible by next-generation DNA sequencing. Mol. Ecol. 2012, 21, 2039–2044. [Google Scholar] [CrossRef]
- Carvalho, D.C. Ichthyoplankton DNA metabarcoding: Challenges and perspectives. Mol. Ecol. 2022, 31, 1612–1614. [Google Scholar] [CrossRef]
- Godinho, A.L.; Lamas, I.R.; Godinho, H.P. Reproductive ecology of Brazilian freshwater fishes. Environ. Biol. Fishes 2010, 87, 143–162. [Google Scholar] [CrossRef]
- Cheshire, K.J.M.; Ye, Q.; Gillanders, B.M.; King, A. Annual variation in larval fish assemblages in a heavily regulated river during differing hydrological conditions. River Res. Appl. 2015, 32, 1207–1219. [Google Scholar] [CrossRef]
- Rosa, G.R.; Salvador, G.N.; Bialetzki, A.; Santos, G.B. Spatial and temporal distribution of ichthyoplankton during an unusual period of low flow in a tributary of the São Francisco River, Brazil. River Res. Appl. 2018, 34, 69–82. [Google Scholar] [CrossRef]
- Soares, M.d.L.; Massaro, M.V.; Hartmann, P.B.; Siveris, S.E.; Pelicice, F.M.; Reynalte-Tataje, D.A. The main channel and river confluences as spawning sites for migratory fishes in the middle Uruguay River. Neotrop. Ichthyol. 2022, 20, e210094. [Google Scholar] [CrossRef]
- Melo-Silva, M.; da Silva, J.C.; Bialetzki, A. Community structure of fish larvae in different biotopes of a neotropical river. Community Ecol. 2022, 23, 1–12. [Google Scholar] [CrossRef]
- CBHSF (Comitê da Bacia Hidrográfica do Rio São Francisco). Diagnóstico da Dimensão Técnica e Institucional Volume 2: Caracterização da Bacia; CBHSF: Belo Horizonte, Brazil, 2016; 292p. [Google Scholar]
- Bazzoli, N. Parâmetros reprodutivos de peixes de interesse comercial na região de Pirapora. In Águas, Peixes e Pescadores do São Francisco das Minas Gerais; PUC Minas: Belo Horizonte, Brazil, 2003; pp. 291–306. [Google Scholar]
- Hermes-Silva, S.; Reynalte-Tataje, D.; Zaniboni-Filho, E. Spatial and temporal distribution of ichthyoplankton in the upper Uruguay River, Brazil. Braz. Arch. Biol. Tech. 2009, 52, 933–944. [Google Scholar] [CrossRef]
- Baumgartner, G.; Nakatani, K.; Gomes, L.; Bialetzki, A.; Sanches, P. Identification of spawning sites and natural nurseries of fishes in the upper Paraná River, Brazil. Environ. Biol. Fishes 2004, 71, 115–125. [Google Scholar] [CrossRef]
- Sato, Y.; Fenerich-Verani, N.; Nuñer, A.P.O.; Godinho, H.P.; Verani, J.R. Padrões reprodutivos de peixes da bacia do São Francisco. In Águas, Peixes e Pescadores do São Francisco das Minas Gerais; PUC Minas: Belo Horizonte, Brazil, 2003; pp. 229–274. [Google Scholar]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Nobile, A.B.; Freitas-Souza, D.; Ruiz-Ruano, F.J.; Nobile, M.L.M.; Costa, G.O.; De Lima, F.P.; Camacho, J.P.M.; Foresti, F.; Oliveira, C. DNA metabarcoding of Neotropical ichthyoplankton: Enabling high accuracy with lower cost. MBMG 2019, 3, e35060. [Google Scholar] [CrossRef]
- Sato, Y.; Godinho, H.P. Migratory Fishes of the São Francisco River. In Migratory Fishes of South America: Biology, Fisheries and Conservation Status; Carolsfeld, J., Harvey, B., Ross, C., Baer, A., Eds.; IDRC-World Bank: Ottawa, ON, Canada, 2003; pp. 195–232. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.; Solymos, P.; Stevens, M.; Wagner, H. Vegan: Community Ecology Package. R Package, Version 2.5.6.1-29; 2015. Available online: https://CRAN.R-project.org/package=vegan (accessed on 1 August 2022).
- Sato, Y.; Cardoso, E.L.; Amorim, J.C.C. Peixes das Lagoas Marginais do Rio São Francisco a Montante da Represa de Três Marias (Minas Gerais), 3rd ed.; CODEVASF: Brasília, Brazil, 1988. [Google Scholar]
- Blumer, L.S. A bibliography and categorization of bony fishes exhibiting parental care. Zool. J. Linn. 1982, 75, 1–22. [Google Scholar] [CrossRef]
- Calegari, B.B.; Vari, R.P.; Reis, R.E. Phylogenetic systematics of the driftwood catfishes (Siluriformes: Auchenipteridae): A combined morphological and molecular analysis. Zool. J. Linn. 2019, 187, 661–773. [Google Scholar] [CrossRef]
- Thibault, R.E.; Schultz, R.J. Reproductive adaptations among viviparous fishes (Cyprinodontiformes: Poeciliidae). Evolution 1978, 32, 320–333. [Google Scholar] [CrossRef]
- Veloso-Junior, V.C.; Arantes, F.P.; dos Santos, J.E. Short communication Population structure and reproduction of the Neotropical catfish Pimelodus pohli, Ribeiro and Lucena, 2006 (Teleostei: Pimelodidae). J. Appl. Ichthyol. 2014, 30, 399–402. [Google Scholar] [CrossRef]
- Reynalte-Tataje, D.A.; Lopes, C.A.; Massaro, M.V.; Hartmann, P.B.; Sulzbacher, R.; Santos, J.A.; Bialetzki, A. State of the art of identification of eggs and larvae of freshwater fish in Brazil. Acta Limnol. Bras. 2020, 32, e6. [Google Scholar] [CrossRef]
- Suzuki, F.M.; Pires, L.V.; Pompeu, P.S. Passage of fish larvae and eggs through the Funil, Itutinga and Camargos Reservoirs on the upper Rio Grande (Minas Gerais, Brazil). Neotrop. Ichthyol. 2011, 9, 617–622. [Google Scholar] [CrossRef]
- Lopes, C.A.; Zaniboni-Filho, E. Mosaic environments shape the distribution of Neotropical freshwater ichthyoplankton. Ecol. Freshw. Fish 2019, 28, 544–553. [Google Scholar] [CrossRef]
- Azevedo-Santos, V.M.; Daga, V.S.; Pelicice, F.M.; Henry, R. Drifting in a free-flowing river: Distribution of fish eggs and larvae in a small tributary of a Neotropical reservoir. Biota Neotrop. 2021, 21, e20211227. [Google Scholar] [CrossRef]
- Loures, R.C. Effectiveness of Fish Monitoring Programs in Hydropower Plant Reservoirs, Lavras. Ph.D. Thesis, Federal University of Lavras, Lavras, Brazil, 2019. [Google Scholar]
- Bialetzki, A.; Tataje, D.; Oliveira, E.; Zaniboni-Filho, E.; Baumgartner, G.; Makrakis, M.; Leite, R. Protocolo mínimo de amostragem do ictioplâncton de água doce para estudos de levantamento, inventário e monitoramento ambiental para implantação de empreendimentos hidrelétricos. Bol. Soc. Bras. Ictiol. 2015, 113, 32–34. [Google Scholar]
- Godinho, A.L.; Kynard, B. Migration and spawning of radio-tagged zulega Prochilodus argenteus in a dammed Brazilian river. Trans. Am. Fish. Soc. 2006, 135, 811–824. [Google Scholar] [CrossRef]
- MMA (Ministério meio Ambiente). Portaria no 148 de 7 de Junho de; Diario Oficial da União: Brasília, Brazil, 2022. [Google Scholar]
- Sato, Y.; Cardoso, E.L.; Godinho, A.L.; Godinho, H.P. Hypophysation parameters of the fish Prochilodus marggravii obtained in routine hatchery station conditions. Rev. Bras. Biol. 1996, 56, 59–64. [Google Scholar]
- Nilsson, C.; Reidy, C.A.; Dynesius, M.; Revenga, C. Fragmentation and flow regulation of the world’s large river systems. Science 2005, 308, 405–408. [Google Scholar] [CrossRef]
- Silva, J.C.; Rosa, R.R.; Galdioli, E.M.; Soares, C.M.; Domingues, W.M.; Veríssimo, S.; Bialetzki, A. Importance of dam-free stretches for fish reproduction: The last remnant in the upper Paraná River. Acta Limnol. Bras. 2017, 29, e106. [Google Scholar] [CrossRef]
- Graça, W.J.; Pavanelli, G.C. Peixes da Planície de Inundação do Alto Rio Paraná e Áreas Adjacentes; Eduem: Maringá, Brazil, 2007; p. 241. [Google Scholar]
- Suzuki, H.I.; Pelicice, F.M.; Luiz, E.A.; Latini, J.D.; Agostinho, A.A. Estratégias reprodutivas da assembléia de peixes da planície de inundação do alto rio Paraná. Pesquisas Ecológicas de Longa Duração–PELD.(Org.). In A Planície Alagável do Rio Paraná: Estrutura e Processos Ambientais; PELD: Maringá, Brazil, 2002. [Google Scholar]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pompeu, P.S.; Wouters, L.; Hilário, H.O.; Loures, R.C.; Peressin, A.; Prado, I.G.; Suzuki, F.M.; Carvalho, D.C. Inadequate Sampling Frequency and Imprecise Taxonomic Identification Mask Results in Studies of Migratory Freshwater Fish Ichthyoplankton. Fishes 2023, 8, 518. https://doi.org/10.3390/fishes8100518
Pompeu PS, Wouters L, Hilário HO, Loures RC, Peressin A, Prado IG, Suzuki FM, Carvalho DC. Inadequate Sampling Frequency and Imprecise Taxonomic Identification Mask Results in Studies of Migratory Freshwater Fish Ichthyoplankton. Fishes. 2023; 8(10):518. https://doi.org/10.3390/fishes8100518
Chicago/Turabian StylePompeu, Paulo Santos, Lídia Wouters, Heron Oliveira Hilário, Raquel Coelho Loures, Alexandre Peressin, Ivo Gavião Prado, Fábio Mineo Suzuki, and Daniel Cardoso Carvalho. 2023. "Inadequate Sampling Frequency and Imprecise Taxonomic Identification Mask Results in Studies of Migratory Freshwater Fish Ichthyoplankton" Fishes 8, no. 10: 518. https://doi.org/10.3390/fishes8100518
APA StylePompeu, P. S., Wouters, L., Hilário, H. O., Loures, R. C., Peressin, A., Prado, I. G., Suzuki, F. M., & Carvalho, D. C. (2023). Inadequate Sampling Frequency and Imprecise Taxonomic Identification Mask Results in Studies of Migratory Freshwater Fish Ichthyoplankton. Fishes, 8(10), 518. https://doi.org/10.3390/fishes8100518