Occurrence of Polyphenols, Isoflavonoids, and Their Metabolites in Milk Samples from Different Cow Feeding Regimens
Abstract
:1. Introduction
2. Materials and Methods
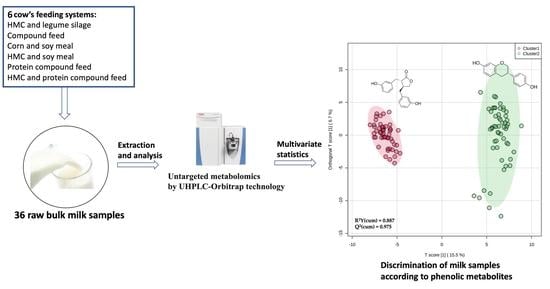
2.1. Collection and Classification of Bulk Milk Samples
2.2. Extraction of Phenolic Compounds
2.3. Phenolic Profiling Based on High-Resolution Mass Spectrometry
2.4. Multivariate Statistical Analysis
3. Results and Discussion
3.1. Profiling of Phenolic Compounds in the Different Milk Samples
3.2. Discrimination of the Milk Samples According to Their Comprehensive Phenolic Profiles
3.3. Discriminative Phenolic Metabolites According to the Supervised OPLS-DA Prediction Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsen, S.Y.; Siew, J.; Lau, E.K.L.; Afiqah Bte Roslee, F.; Chan, H.M.; Loke, W.M. Cow’s milk as a dietary source of equol and phenolic antioxidants: Differential distribution in the milk aqueous and lipid fractions. Dairy Sci. Technol. 2014, 94, 625–632. [Google Scholar] [CrossRef]
- O’Connell, J.E.; Fox, P.F. Significance and applications of phenolic compounds in the production and quality of milk and dairy products: A review. Int. Dairy J. 2001, 11, 103–120. [Google Scholar] [CrossRef]
- Schwen, R.J.; Nguyen, L.; Jackson, R.L. Elucidation of the metabolic pathway of S-equol in rat, monkey and man. Food Chem. Toxicol. 2012, 50, 2074–2083. [Google Scholar] [CrossRef] [PubMed]
- Mayo, B.; Vázquez, L.; Flórez, A.B. Equol: A Bacterial Metabolite from The Daidzein Isoflavone and Its Presumed Beneficial Health Effects. Nutrients 2019, 11, 2231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardana, C.; Canzi, E.; Simonetti, P. R(-)-O-desmethylangolensin is the main enantiomeric form of daidzein metabolite produced by human in vitro and in vivo. J. Chrom. B 2014, 953, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Besle, J.M.; Viala, D.; Martin, B.; Pradel, P.; Meunier, B.; Berdagué, J.L.; Fraisse, D.; Lamaison, J.L.; Coulon, J.B. Ultraviolet-absorbing compounds in milk are related to forage polyphenols. J. Dairy Sci. 2010, 93, 2846–2856. [Google Scholar] [CrossRef] [Green Version]
- Carpio, A.; Bonilla-Valverde, D.; Arce, C.; Rodríguez-Estévez, V.; Sánchez-Rodríguez, M.; Arce, L.; Valcárcel, M. Evaluation of hippuric acid content in goat milk as a marker of feeding regimen. J. Dairy Sci. 2013, 96, 5426–5434. [Google Scholar] [CrossRef] [Green Version]
- Vázquez, C.V.; Rojas, M.G.V.; Ramírez, C.A.; Chávez-Servín, J.L.; García-Gasca, T.; Martínez, R.A.F.; García, O.P.; Rosado, J.L.; López-Sabater, C.M.; Castellote, A.I.; et al. Total phenolic compounds in milk from different species. Design of an extraction technique for quantification using the Folin–Ciocalteu method. Food Chem. 2015, 176, 480–486. [Google Scholar] [CrossRef]
- Gallo, A.; Valsecchi, C.; Masseroni, M.; Cannas, A.; Ghilardelli, F.; Masoero, F.; Atzori, A.S. An observational study to verify the influence of different nutritional corn silage-based strategies on efficient use of dietary nutrients, faecal fermentation profile, and profitability in a cohort of intensive dairy farms. Ital. J. Anim. Sci. 2022, 21, 228–243. [Google Scholar] [CrossRef]
- Rocchetti, G.; Gallo, A.; Nocetti, M.; Lucini, L.; Masoero, F. Milk metabolomics based on ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry to discriminate different cows feeding regimens. Food Res. Int. 2020, 134, 109279. [Google Scholar] [CrossRef]
- Rocchetti, G.; Ghilardelli, F.; Masoero, F.; Gallo, A. Screening of Regulated and Emerging Mycotoxins in Bulk Milk Samples by High-Resolution Mass Spectrometry. Foods 2021, 10, 2025. [Google Scholar] [CrossRef]
- Rocchetti, G.; Ghilardelli, F.; Bonini, P.; Lucini, L.; Masoero, F.; Gallo, A. Changes of Milk Metabolomic Profiles Resulting from a Mycotoxins-Contaminated Corn Silage Intake by Dairy Cows. Metabolites 2021, 11, 475. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Pimpão, R.C.; Ventura, M.R.; Ferreira, R.B.; Williamson, G.; Santos, C.N. Phenolic sulfates as new and highly abundant metabolites in human plasma after ingestion of a mixed berry fruit purée. Br. J. Nutr. 2015, 113, 454–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasta, V.; Daghio, M.; Cappucci, A.; Buccioni, A.; Serra, A.; Viti, C.; Mele, M. Invited review: Plant polyphenols and rumen microbiota responsible for fatty acid biohydrogenation, fiber digestion, and methane emission: Experimental evidence and methodological approaches. J. Dairy Sci. 2019, 102, 3781–3804. [Google Scholar] [CrossRef] [PubMed]
- Hook, S.E.; Wright, A.D.G.; McBride, B.W. Methanogens: Methane producers of the rumen and mitigation strategies. Archaea 2010, 2010, 945785. [Google Scholar] [CrossRef] [Green Version]
- Hashem, N.M.; Gonzalez-Bulnes, A.; Simal-Gandara, J. Polyphenols in Farm Animals: Source of Reproductive Gain or Waste? Antioxidants 2020, 9, 1023. [Google Scholar] [CrossRef]
- Brito, A.F.; Zang, Y. A Review of Lignan Metabolism, Milk Enterolactone Concentration, and Antioxidant Status of Dairy Cows Fed Flaxseed. Molecules 2019, 24, 41. [Google Scholar] [CrossRef] [Green Version]
- Senizza, A.; Rocchetti, G.; Mosele, J.I.; Patrone, V.; Callegari, M.L.; Morelli, L.; Lucini, L. Lignans and Gut Microbiota: An Interplay Revealing Potential Health Implications. Molecules 2020, 25, 5709. [Google Scholar] [CrossRef]
- Peñalvo, J.L.; Heinonen, S.M.; Nurmi, T.; Deyama, T.; Nishibe, S.; Adlercreutz, H. Plant Lignans in Soy-Based Health Supplements. J. Agr. Food Chem. 2004, 52, 4133–4138. [Google Scholar] [CrossRef]
- Côrtes, C.; Gagnon, N.; Benchaar, C.; da Silva, D.; Santos, G.T.D.; Petit, H.V. In vitro metabolism of flax lignans by ruminal and faecal microbiota of dairy cows. J. Appl. Microbiol. 2008, 105, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Petit, H.V.; Gagnon, N. Milk concentrations of the mammalian lignans enterolactone and enterodiol, milk production, and whole tract digestibility of dairy cows fed diets containing different concentrations of flaxseed meal. Anim. Feed Sci. Technol. 2009, 152, 103–111. [Google Scholar] [CrossRef]
- Křížová, L.; Křešt’áková, V.; Dadáková, K.; Kašparovský, T. Production of Bovine Equol-Enriched Milk: A Review. Animals 2021, 11, 735. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.J.; Griffin, C.; Rai, D.K.; O’Callaghan, T.F.; O’Sullivan, M.G.; Kerry, J.P.; Kilcawley, K.N. Dietary Compounds Influencing the Sensorial, Volatile and Phytochemical Properties of Bovine Milk. Molecules 2020, 25, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wójciak–Kosior, M.; Dresler, S.; Sowa, I.; Łuć, K.; Staniak, M.; Latalski, M.; Kiełbowicz, K.Z.; Kocjan, R. Effect of various strontium concentrations on its uptake and the content of isoflavonesin soybean sprouts. Acta Biolog. Crac. Ser. Bot. 2019, 61, 7–12. [Google Scholar]
- Chiriac, E.R.; Chiţescu, C.L.; Borda, D.; Lupoae, M.; Gird, C.E.; Geană, E.-I.; Blaga, G.-V.; Boscencu, R. Comparison of the Polyphenolic Profile of Medicago sativa L. and Trifolium pratense L. Sprouts in Different Germination Stages Using the UHPLC-Q Exactive Hybrid Quadrupole Orbitrap High-Resolution Mass Spectrometry. Molecules 2020, 25, 2321. [Google Scholar] [CrossRef] [PubMed]
- Bláhová, L.; Kohoutek, J.; Procházková, T.; Prudíková, M.; Bláha, L. Phytoestrogens in milk: Overestimations caused by contamination of the hydrolytic enzyme used during sample extraction. J. Dairy Sci. 2016, 99, 6973–6982. [Google Scholar] [CrossRef]
- Wocławek-Potocka, I.; Mannelli, C.; Boruszewska, D.; Kowalczyk-Zieba, I.; Waśniewski, T.; Skarzyński, D.J. Diverse effects of phytoestrogens on the reproductive performance: Cow as a model. Int. J. Endocrinol. 2013, 2013, 650984. [Google Scholar] [CrossRef] [Green Version]
- Patton, S. The Presence of Hippuric Acid in Milk. J. Dairy Sci. 1953, 36, 943–947. [Google Scholar] [CrossRef]
- Carpio, A.; Rodríguez-Estévez, V.; Sánchez-Rodríguez, M.; Arce, L.; Valcárcel, M. Differentiation of organic goat’s milk based on its hippuric acid content as determined by capillary electrophoresis. Electrophoresis 2010, 31, 2211–2217. [Google Scholar] [CrossRef]
- bin Cao, B.; Jin, X.; Yang, H.J.; Li, S.L.; Jiang, L.S. Microbial release of ferulic and p-coumaric acids from forages and their digestibility in lactating cows fed total mixed rations with different forage combinations. J. Sci. Food Agric. 2016, 96, 650–655. [Google Scholar]
- Stefanello, F.S.; Santos, C.O.; Bochi, V.C.; Fruet, A.P.B.; Soquetta, M.B.; Dörr, A.C.; Nörnberg, J.L. Analysis of polyphenols in brewer’s spent grain and its comparison with corn silage and cereal brans commonly used for animal nutrition. Food Chem. 2018, 239, 385–401. [Google Scholar] [CrossRef] [PubMed]


| Feeding Cluster * | Feeding Regimen | Milk Sample (ID) | Phenolic Cluster ** |
|---|---|---|---|
| CL1 | HMC and legume silage strategy | Sample 4 | Cluster 2 |
| − | Sample 15 | Cluster 1 | |
| − | Sample 34 | Cluster 1 | |
| CL2 | Compound feed strategy | Sample 20 | Cluster 2 |
| Sample 33 | Cluster 1 | ||
| CL3 | Corn and soy meals strategy | Sample 5 | Cluster 2 |
| − | Sample 6 | Cluster 2 | |
| − | Sample 9 | Cluster 1 | |
| − | Sample 19 | Cluster 1 | |
| − | Sample 22 | Cluster 1 | |
| − | Sample 23 | Cluster 2 | |
| − | Sample 24 | Cluster 1 | |
| − | Sample 36 | Cluster 1 | |
| CL4 | HMC and soy meal strategy | Sample 1 | Cluster 2 |
| − | Sample 3 | Cluster 2 | |
| − | Sample 8 | Cluster 1 | |
| − | Sample 11 | Cluster 2 | |
| − | Sample 12 | Cluster 1 | |
| − | Sample 17 | Cluster 1 | |
| − | Sample 18 | Cluster 1 | |
| − | Sample 21 | Cluster 2 | |
| − | Sample 25 | Cluster 1 | |
| − | Sample 27 | Cluster 2 | |
| − | Sample 28 | Cluster 2 | |
| − | Sample 29 | Cluster 1 | |
| − | Sample 31 | Cluster 1 | |
| CL5 | Protein compound feed strategy | Sample 2 | Cluster 2 |
| − | Sample 10 | Cluster 2 | |
| − | Sample 13 | Cluster 2 | |
| − | Sample 16 | Cluster 2 | |
| − | Sample 26 | Cluster 2 | |
| − | Sample 32 | Cluster 2 | |
| − | Sample 37 | Cluster 1 | |
| CL6 | HMC and protein compound feeds strategy | Sample 7 | Cluster 1 |
| − | Sample 14 | Cluster 2 | |
| − | Sample 30 | Cluster 1 |
| Discriminant Metabolites | Phenolic Subclass | VIP Score (OPLS-DA) | Log2 Fold-Change “Cluster 1 vs. Cluster 2” | p-Value (ANOVA, FDR) |
|---|---|---|---|---|
| Enterolactone | Lignans | 2.290 | 3.58 | 7.9 × 10−36 |
| p-Anisaldehyde | Hydroxybenzaldehydes | 2.271 | 3.05 | 8.5 × 10−35 |
| 4′,7-Dihydroxy-3′-methoxyisoflavan | Isoflavonoids | 2.287 | −5.39 | 3.5 × 10−34 |
| Quercetin 3-O-xylosyl-glucuronide | Flavonols | 2.254 | 3.42 | 7.85 × 10−34 |
| Vanillic acid | Hydroxybenzoic acids | 2.238 | 2.83 | 2.14 × 10−33 |
| Gardenin B | Flavones | 2.217 | −6.50 | 6.71 × 10−31 |
| Piceatannol | Stilbenes | 2.199 | −4.35 | 8.84 × 10−31 |
| Ellagic acid | Hydroxybenzoic acids | 2.236 | −1.38 | 1.14 × 10−30 |
| 7-Hydroxysecoisolariciresinol | Lignans | 2.161 | −4.19 | 1.28 × 10−28 |
| O-Desmethylangolensin | Isoflavonoids | 2.147 | −5.02 | 7.83 × 10−28 |
| 4′-Hydroxy-3,4,5-trimethoxystilbene | Stilbenes | 2.132 | −4.98 | 2.51 × 10−26 |
| Hesperetin 3′,7-O-diglucuronide | Flavanones | 1.967 | −12.23 | 3.15 × 10−20 |
| 6″-O-Malonylgenistin | Isoflavonoids | 1.805 | −1.84 | 2.23 × 10−16 |
| Cinnamic acid | Hydroxycinnamic acids | 1.773 | −1.38 | 1.57 × 10−15 |
| Homoveratric acid | Hydroxyphenylacetic acids | 1.608 | −1.13 | 1.52 × 10−12 |
| Equol | Isoflavonoids | 1.588 | −1.60 | 1.39 × 10−10 |
| 3-Caffeoylquinic acid | Hydroxycinnamic acids | 1.398 | 1.09 | 8.51 × 10−9 |
| Formononetin | Isoflavonoids | 1.318 | 0.77 | 7.02 × 10−8 |
| Hippuric acid | Hydroxybenzoic acids | 1.176 | 0.56 | 7.97 × 10−7 |
| 4-Hydroxyhippuric acid | Hydroxybenzoic acids | 1.162 | 1.03 | 1.44 × 10−6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocchetti, G.; Ghilardelli, F.; Mosconi, M.; Masoero, F.; Gallo, A. Occurrence of Polyphenols, Isoflavonoids, and Their Metabolites in Milk Samples from Different Cow Feeding Regimens. Dairy 2022, 3, 314-325. https://doi.org/10.3390/dairy3020024
Rocchetti G, Ghilardelli F, Mosconi M, Masoero F, Gallo A. Occurrence of Polyphenols, Isoflavonoids, and Their Metabolites in Milk Samples from Different Cow Feeding Regimens. Dairy. 2022; 3(2):314-325. https://doi.org/10.3390/dairy3020024
Chicago/Turabian StyleRocchetti, Gabriele, Francesca Ghilardelli, Martina Mosconi, Francesco Masoero, and Antonio Gallo. 2022. "Occurrence of Polyphenols, Isoflavonoids, and Their Metabolites in Milk Samples from Different Cow Feeding Regimens" Dairy 3, no. 2: 314-325. https://doi.org/10.3390/dairy3020024
APA StyleRocchetti, G., Ghilardelli, F., Mosconi, M., Masoero, F., & Gallo, A. (2022). Occurrence of Polyphenols, Isoflavonoids, and Their Metabolites in Milk Samples from Different Cow Feeding Regimens. Dairy, 3(2), 314-325. https://doi.org/10.3390/dairy3020024