Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen
Abstract
:1. Introduction
2. Materials and Methods
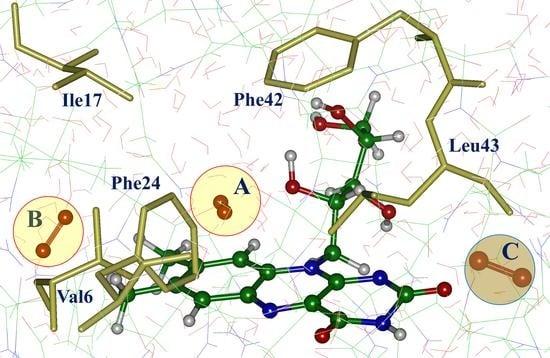
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Juan, C.A.; Pérez de la Lastra, J.M.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef] [PubMed]
- Romero, E.; Gómez Castellanos, J.R.; Gadda, G.; Fraaije, M.W.; Mattevi, A. Same substrate, many reactions: Oxygen activation in flavoenzymes. Chem. Rev. 2018, 118, 1742–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, X.; Lev-Ram, V.; Deerinck, T.J.; Qi, Y.; Ramko, E.B.; Davidson, M.W.; Jin, Y.; Ellisman, M.H.; Tsien, R.Y. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011, 9, e1001041. [Google Scholar] [CrossRef] [Green Version]
- Torra, J.; Lafaye, C.; Signor, L.; Aumonier, S.; Flors, C.; Shu, X.; Nonell, S.; Gotthard, G.; Royant, A. Tailing miniSOG: Structural bases of the complex photophysics of a favin-binding singlet oxygen photosensitizing protein. Sci. Rep. 2019, 9, 2428. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Lafaye, C.; Aumonier, S.; Torra, J.; Signor, L.; von Stetten, D.; Noirclerc-Savoye, M.; Shu, X.; Ruiz-González, R.; Gotthard, G.; Royant, A.; et al. Ribofavin-binding proteins for singlet oxygen production. Photochem. Photobiol. Sci. 2022, 21, 1545–1555. [Google Scholar] [CrossRef] [PubMed]
- Colloc’h, N.; Gabison, L.; Monard, G.; Altarsha, M.; Chiadmi, M.; Marassio, G.; Sopkova-de Oliveira Santos, J.; El Hajji, M.; Castro, B.; Abraini, J.H.; et al. Oxygen pressurized X-ray crystallography: Probing the dioxygen binding site in cofactorless urate oxidase and implications for its catalytic mechanism. Biophys. J. 2008, 95, 2415–2422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, A.; Saleem-Batcha, R.; Sanders, J.N.; Stull, F.; Houk, K.N.; Teufel, R. Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases. Nat. Chem. Biol. 2020, 16, 556–563. [Google Scholar] [CrossRef]
- Auhim, H.S.; Grigorenko, B.L.; Harris, T.K.; Aksakal, O.E.; Polyakov, I.V.; Berry, C.; Gomes, G.d.P.; Alabugin, I.V.; Rizkallah, P.J.; Nemukhin, A.V.; et al. Stalling chromophore synthesis of the fluorescent protein venus reveals the molecular basis of the final oxidation step. Chem. Sci. 2021, 12, 7735–7745. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.; Kim, K.; King, P.; Seibert, M.; Schulten, K. Finding Gas Diffusion Pathways in Proteins: Application to O2 and H2 Transport in CpI [FeFe]-Hydrogenase and the Role of Packing Defects. Structure 2005, 13, 1321–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Carpentier, P.; Bourgeois, D.; Field, M. Diffusion Pathways of Oxygen Species in the Phototoxic Fluorescent Protein KillerRed. Photochem. Photobiol. Sci. 2010, 9, 1342–1350. [Google Scholar] [CrossRef]
- Chapagain, P.P.; Regmi, C.K.; Castillo, W. Fluorescent Protein Barrel Fluctuations and Oxygen Diffusion Pathways in mCherry. J. Chem. Phys. 2011, 135, 235101. [Google Scholar] [CrossRef] [Green Version]
- Regmi, C.K.; Bhandari, Y.R.; Gerstman, B.S.; Chapagain, P.P. Exploring the Diffusion of Molecular Oxygen in the Red Fluorescent Protein mCherry Using Explicit Oxygen Molecular Dynamics Simulations. J. Phys. Chem. B 2013, 117, 2247–2253. [Google Scholar] [CrossRef] [Green Version]
- Polyakov, I.V.; Domratcheva, T.M.; Kulakova, A.M.; Nemukhin, A.V.; Grigorenko, B.L. Computational Modeling of the Interaction of Molecular Oxygen with the Flavin-Dependent Enzyme RutA. Supercomput. Front. Innov. 2022, 9, 46–55. [Google Scholar] [CrossRef]
- Polyakov, I.V.; Nemukhin, A.V.; Domratcheva, T.M.; Kulakova, A.M.; Grigorenko, B.L. Quantum-based modeling of protein-ligand interaction: The complex of RutA with uracil and molecular oxygen. Mol. Inform. 2022, 41, 2200175. [Google Scholar] [CrossRef] [PubMed]
- Grigorenko, B.L.; Nemukhin, A.V.; Polyakov, I.V.; Khrenova, M.G.; Krylov, A.I. A light-induced reaction with oxygen leads to chromophore decomposition and irreversible photobleaching in GFP-type proteins. J. Phys. Chem. B 2015, 119, 5444–5452. [Google Scholar] [CrossRef]
- Grigorenko, B.L.; Krylov, A.I.; Nemukhin, A.V. Molecular Modeling Clarifies the Mechanism of Chromophore Maturation in the Green Fluorescent Protein. J. Am. Chem. Soc. 2017, 139, 10239–10249. [Google Scholar] [CrossRef]
- Grigorenko, B.L.; Domratcheva, T.M.; Nemukhin, A.V. QM/MM Modeling of the Flavin Functionalization in the RutA Monooxygenase. Molecules 2023, 28, 2405. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [Green Version]
- Aleksandrov, A. A Molecular Mechanics Model for Flavins. J. Comp. Chem. 2019, 40, 2834–2842. [Google Scholar] [CrossRef]
- Wang, S.; Hou, K.; Heinz, H. Accurate and Compatible Force Fields for Molecular Oxygen, Nitrogen, and Hydrogen to Simulate Gases, Electrolytes, and Heterogeneous Interfaces. J. Chem. Theory Comput. 2021, 17, 5198–5213. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Chai, J.-D.; Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef] [Green Version]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Melo, M.C.R.; Bernardi, R.C.; Rudack, T.; Scheurer, M.; Riplinger, C.; Phillips, J.C.; Maia, J.D.C.; Rocha, G.B.; Ribeiro, J.V.; Stone, J.E.; et al. NAMD Goes Quantum: An Integrative Suite for Hybrid Simulations. Nat. Methods 2018, 15, 351–354. [Google Scholar] [CrossRef]
- Seritan, S.; Bannwarth, C.; Fales, B.S.; Hohenstein, E.G.; Isborn, C.M.; Kokkila-Schumacher, S.I.L.; Li, X.; Liu, F.; Luehr, N.; Snyder, J.W., Jr.; et al. Terachem: A graphical processing unit-accelerated electronic structure package for large-scale ab initio molecular dynamics. WIREs Comput. Mol. Sci. 2021, 11, e1494. [Google Scholar] [CrossRef]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef] [Green Version]








| Model System | BB 5–105 | BB 28–48 | 28–48 noH | ISO |
|---|---|---|---|---|
| miniSOG[FMN] | 0.8 ± 0.2 | 0.7 ± 0.2 | 1.4 ± 0.3 | 0.9 ± 0.2 |
| miniSOG[RF] | 1.1 ± 0.2 | 1.1 ± 0.3 | 2.0 ± 0.3 | 1.0 ± 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polyakov, I.; Kulakova, A.; Nemukhin, A. Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen. Biophysica 2023, 3, 252-262. https://doi.org/10.3390/biophysica3020016
Polyakov I, Kulakova A, Nemukhin A. Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen. Biophysica. 2023; 3(2):252-262. https://doi.org/10.3390/biophysica3020016
Chicago/Turabian StylePolyakov, Igor, Anna Kulakova, and Alexander Nemukhin. 2023. "Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen" Biophysica 3, no. 2: 252-262. https://doi.org/10.3390/biophysica3020016
APA StylePolyakov, I., Kulakova, A., & Nemukhin, A. (2023). Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen. Biophysica, 3(2), 252-262. https://doi.org/10.3390/biophysica3020016