Molecular Tools for Diagnosis and Surveillance of Soil-Transmitted Helminths in Endemic Areas
Abstract
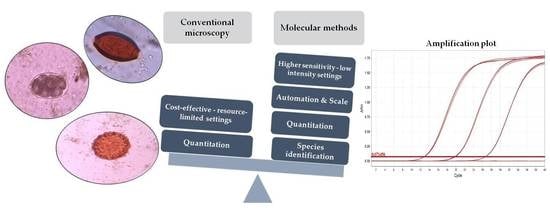
:1. Introduction
2. Role of Molecular Methods
3. Sample Storage and DNA Extraction Methods
4. Conventional PCR
5. Real-Time PCR
6. Isothermal Assay (LAMP)
7. Other Recent Technical Developments
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pullan, R.L.; Smith, J.L.; Jasrasaria, R.; Brooker, S.J. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit Vectors 2014, 7, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nokes, C.; Grantham-McGregor, S.M.; Sawyer, A.W.; Cooper, E.S.; Robinson, B.A.; Bundy, D.A. Moderate to heavy infections of Trichuris trichiura affect cognitive function in Jamaican school children. Parasitology 1992, 104, 539–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nokes, C.; Bundy, D.A. Compliance and absenteeism in school children: Implications for helminth control. Trans. R. Soc. Trop. Med. Hyg. 1993, 87, 148–152. [Google Scholar] [CrossRef]
- Savioli, L.; Bundy, D.; Tomkins, A. Intestinal parasitic infections: A soluble public health problem. Trans. R. Soc. Trop. Med. Hyg. 1992, 86, 353–354. [Google Scholar] [CrossRef] [Green Version]
- WHO. 2030 Targets for Soil-Transmitted Helminthiases Control Programmes; World Health Organization: Geneva, Switzerland, 2020; Available online: https://apps.who.int/iris/handle/10665/330611 (accessed on 8 July 2020).
- WHO. Assessing the Epidemiology of Soil-Transmitted Helminths during a Transmission Assessment Survey (TAS); World Health Organization: Geneva, Switzerland, 2015; Available online: http://www.who.int/intestinal_worms/resources/9789241508384/en/ (accessed on 11 July 2020).
- Montresor, A.; WHO. Helminth Control in School Age Children; World Health Organization: Geneva, Switzerland, 2002; Available online: http://www.who.int/intestinal_worms/resources/9789241548267/en/ (accessed on 8 July 2020).
- Mengist, H.M.; Demeke, G.; Zewdie, O.; Belew, A. Diagnostic performance of direct wet mount microscopy in detecting intestinal helminths among pregnant women attending ante-natal care (ANC) in East Wollega, Oromia, Ethiopia. BMC Res. Notes 2018, 11, 276. [Google Scholar] [CrossRef] [Green Version]
- Endris, M.; Tekeste, Z.; Lemma, W.; Kassu, A. Comparison of the Kato-Katz, Wet Mount, and Formol-Ether Concentration Diagnostic Techniques for Intestinal Helminth Infections in Ethiopia. ISRN Parasitol. 2013, 2013, 180439. [Google Scholar] [CrossRef] [Green Version]
- Cools, P.; Vlaminck, J.; Albonico, M.; Ame, S.; Ayana, M.; José Antonio, B.P.; Cringoli, G.; Dana, D.; Keiser, J.; Maurelli, M.P.; et al. Diagnostic performance of a single and duplicate Kato-Katz, Mini-FLOTAC, FECPAKG2 and qPCR for the detection and quantification of soil-transmitted helminths in three endemic countries. PLoS Negl. Trop. Dis. 2019, 13, e0007446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarafder, M.R.; Carabin, H.; Joseph, L.; Balolong, E.; Olveda, R.; McGarvey, S.T. Estimating the sensitivity and specificity of Kato-Katz stool examination technique for detection of hookworms, Ascaris lumbricoides and Trichuris trichiura infections in humans in the absence of a “gold standard”. Int. J. Parasitol. 2010, 40, 399–404. [Google Scholar] [CrossRef] [Green Version]
- Levecke, B.; Behnke, J.M.; Ajjampur, S.S.; Albonico, M.; Ame, S.M.; Charlier, J.; Geiger, S.M.; Hoa, N.T.; Kamwa Ngassam, R.I.; Kotze, A.C.; et al. A comparison of the sensitivity and fecal egg counts of the McMaster egg counting and Kato-Katz thick smear methods for soil-transmitted helminths. PLoS Negl. Trop. Dis. 2011, 5, e1201. [Google Scholar] [CrossRef] [Green Version]
- Cringoli, G.; Rinaldi, L.; Maurelli, M.P.; Utzinger, J. FLOTAC: New multivalent techniques for qualitative and quantitative copromicroscopic diagnosis of parasites in animals and humans. Nat. Protoc. 2010, 5, 503–515. [Google Scholar] [CrossRef]
- Habtamu, K.; Degarege, A.; Ye-Ebiyo, Y.; Erko, B. Comparison of the Kato-Katz and FLOTAC techniques for the diagnosis of soil-transmitted helminth infections. Parasitol. Int. 2011, 60, 398–402. [Google Scholar] [CrossRef]
- Knopp, S.; Speich, B.; Hattendorf, J.; Rinaldi, L.; Mohammed, K.A.; Khamis, I.S.; Mohammed, A.S.; Albonico, M.; Rollinson, D.; Marti, H.; et al. Diagnostic Accuracy of Kato-Katz and FLOTAC for Assessing Anthelmintic Drug Efficacy. PLoS Negl. Trop. Dis. 2011, 5, e1036. [Google Scholar] [CrossRef] [PubMed]
- Nikolay, B.; Brooker, S.J.; Pullan, R.L. Sensitivity of diagnostic tests for human soil-transmitted helminth infections: A meta-analysis in the absence of a true gold standard. Int. J. Parasitol. 2014, 44, 765–774. [Google Scholar] [CrossRef] [Green Version]
- Moser, W.; Bärenbold, O.; Mirams, G.J.; Cools, P.; Vlaminck, J.; Ali, S.M.; Ame, S.M.; Hattendorf, J.; Vounatsou, P.; Levecke, B.; et al. Diagnostic comparison between FECPAKG2 and the Kato-Katz method for analyzing soil-transmitted helminth eggs in stool. PLoS Negl. Trop. Dis. 2018, 12, e0006562. [Google Scholar] [CrossRef] [PubMed]
- Sukas, S.; Van Dorst, B.; Kryj, A.; Lagatie, O.; De Malsche, W.; Stuyver, L.J. Development of a Lab-on-a-Disk Platform with Digital Imaging for Identification and Counting of Parasite Eggs in Human and Animal Stool. Micromachines 2019, 10, 852. [Google Scholar] [CrossRef] [Green Version]
- Yang, A.; Bakhtari, N.; Langdon-Embry, L.; Redwood, E.; Grandjean Lapierre, S.; Rakotomanga, P.; Rafalimanantsoa, A.; De Dios Santos, J.; Vigan-Womas, I.; Knoblauch, A.M.; et al. Kankanet: An artificial neural network-based object detection smartphone application and mobile microscope as a point-of-care diagnostic aid for soil-transmitted helminthiases. PLoS Negl. Trop. Dis. 2019, 13, e0007577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmström, O.; Linder, N.; Ngasala, B.; Mårtensson, A.; Linder, E.; Lundin, M.; Moilanen, H.; Suutala, A.; Diwan, V.; Lundin, J. Point-of-care mobile digital microscopy and deep learning for the detection of soil-transmitted helminths and Schistosoma haematobium. Glob. Health Action 2017, 10 (Suppl. S3), 1337325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Periago, M.V.; Diniz, R.C.; Pinto, S.A.; Yakovleva, A.; Correa-Oliveira, R.; Diemert, D.J.; Bethony, J.M. The Right Tool for the Job: Detection of Soil-Transmitted Helminths in Areas Co-endemic for Other Helminths. PLoS Negl. Trop. Dis. 2015, 9, e0003967. [Google Scholar] [CrossRef]
- Albonico, M.; Rinaldi, L.; Sciascia, S.; Morgoglione, M.E.; Piemonte, M.; Maurelli, M.P.; Musella, V.; Utzinger, J.; Ali, S.M.; Ame, S.M.; et al. Comparison of three copromicroscopic methods to assess albendazole efficacy against soil-transmitted helminth infections in school-aged children on Pemba Island. Trans. R. Soc. Trop. Med. Hyg. 2013, 107, 493–501. [Google Scholar] [CrossRef] [Green Version]
- Barda, B.; Cajal, P.; Villagran, E.; Cimino, R.; Juarez, M.; Krolewiecki, A.; Rinaldi, L.; Cringoli, G.; Burioni, R.; Albonico, M. Mini-FLOTAC, Kato-Katz and McMaster: Three methods, one goal; highlights from north Argentina. Parasit. Vectors 2014, 7, 271. [Google Scholar] [CrossRef] [Green Version]
- Albonico, M.; Ame, S.M.; Vercruysse, J.; Levecke, B. Comparison of the Kato-Katz thick smear and McMaster egg counting techniques for monitoring drug efficacy against soil-transmitted helminths in schoolchildren on Pemba Island, Tanzania. Trans. R. Soc. Trop. Med. Hyg. 2012, 106, 199–201. [Google Scholar] [CrossRef]
- Bärenbold, O.; Raso, G.; Coulibaly, J.T.; N’Goran, E.K.; Utzinger, J.; Vounatsou, P. Estimating sensitivity of the Kato-Katz technique for the diagnosis of Schistosoma mansoni and hookworm in relation to infection intensity. PLoS Negl. Trop. Dis. 2017, 11, e0005953. [Google Scholar] [CrossRef]
- Liu, C.; Lu, L.; Zhang, L.; Bai, Y.; Medina, A.; Rozelle, S.; Smith, D.S.; Zhou, C.; Zang, W. More Poop, More Precision: Improving Epidemiologic Surveillance of Soil-Transmitted Helminths with Multiple Fecal Sampling using the Kato–Katz Technique. Am. J. Trop. Med. Hyg. 2017, 97, 870–875. [Google Scholar] [CrossRef]
- Knopp, S.; Salim, N.; Schindler, T.; Voules, D.A.K.; Rothen, J.; Lweno, O.; Mohammed, A.S.; Singo, R.; Benninghoff, M.; Nsojo, A.A.; et al. Diagnostic Accuracy of Kato–Katz, FLOTAC, Baermann, and PCR Methods for the Detection of Light-Intensity Hookworm and Strongyloides stercoralis Infections in Tanzania. Am. J. Trop. Med. Hyg. 2014, 90, 535–545. [Google Scholar] [CrossRef] [Green Version]
- Easton, A.V.; Oliveira, R.G.; O’Connell, E.M.; Kepha, S.; Mwandawiro, C.S.; Njenga, S.M.; Kihara, J.H.; Mwatele, C.; Odiere, M.R.; Brooker, S.J.; et al. Multi-parallel qPCR provides increased sensitivity and diagnostic breadth for gastrointestinal parasites of humans: Field-based inferences on the impact of mass deworming. Parasit. Vectors 2016, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Benjamin-Chung, J.; Pilotte, N.; Ercumen, A.; Grant, J.R.; Maasch, J.R.; Gonzalez, A.M.; Ester, A.C.; Arnold, B.F.; Rahman, M.; Haque, R.; et al. Comparison of multi-parallel qPCR and double-slide Kato-Katz for detection of soil-transmitted helminth infection among children in rural Bangladesh. PLoS Negl. Trop. Dis. 2020, 14, e0008087. [Google Scholar] [CrossRef]
- Turner, H.C.; Bettis, A.A.; Dunn, J.C.; Whitton, J.M.; Hollingsworth, T.D.; Fleming, F.M.; Anderson, R.M. Economic Considerations for Moving beyond the Kato-Katz Technique for Diagnosing Intestinal Parasites as We Move Towards Elimination. Trends Parasitol. 2017, 33, 435–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ásbjörnsdóttir, K.H.; Ajjampur, S.S.R.; Anderson, R.M.; Bailey, R.; Gardiner, I.; Halliday, K.E.; Ibikounle, M.; Kalua, K.; Kang, G.; Littlewood, D.T.J.; et al. Assessing the feasibility of interrupting the transmission of soil-transmitted helminths through mass drug administration: The DeWorm3 cluster randomized trial protocol. PLoS Negl. Trop. Dis. 2018, 12, e0006166. [Google Scholar] [CrossRef] [PubMed]
- Brooker, S.J.; Mwandawiro, C.S.; Halliday, K.E.; Njenga, S.M.; Mcharo, C.; Gichuki, P.M.; Wasunna, B.; Kihara, J.H.; Njomo, D.; Alusala, D.; et al. Interrupting transmission of soil-transmitted helminths: A study protocol for cluster randomised trials evaluating alternative treatment strategies and delivery systems in Kenya. BMJ Open 2015, 5, e008950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The DeWorm3 Project Team; Hasegawa, M.; Pilotte, N.; Kikuchi, M.; Means, A.R.; Papaiakovou, M.; Gonzalez, A.M.; Maasch, J.R.; Ikuno, H.; Sunahara, T.; et al. What does soil-transmitted helminth elimination look like? Results from a targeted molecular detection survey in Japan. Parasit. Vectors 2020, 13, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verweij, J.J.; Brienen, E.A.T.; Ziem, J.; Polderman, A.M.; Yelifari, L.; Van Lieshout, L. Simultaneous Detection and Quantification of Ancylostoma duodenale, Necator americanus, and Oesophagostomum bifurcum in Fecal Samples Using Multiplex Real-Time PCR. Am. J. Trop. Med. Hyg. 2007, 77, 685–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilotte, N.; Papaiakovou, M.; Grant, J.R.; Bierwert, L.A.; Llewellyn, S.; McCarthy, J.S.; Williams, S.A. Improved PCR-Based Detection of Soil Transmitted Helminth Infections Using a Next-Generation Sequencing Approach to Assay Design. PLoS Negl. Trop. Dis. 2016, 10, e0004578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, B.; Li, T.; Xiao, S.; Zheng, F.; Hawdon, J.M. Species-Specific Identification of Human Hookworms by PCR of the Mitochondrial Cytochrome Oxidase I Gene. J. Parasitol. 2001, 87, 1227. [Google Scholar] [CrossRef]
- Fairley, T.L.; Kilpatrick, C.W.; Conn, J.E. Intragenomic heterogeneity of internal transcribed spacer rDNA in neotropical malaria vector Anopheles aquasalis (Diptera: Culicidae). J. Med. Entomol. 2005, 42, 795–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.; Singh, M.N.; Bera, A.K.; Bandyopadhyay, S.; Bhattacharya, D. Molecular basis for identification of species/isolates of gastrointestinal nematode parasites. Asian Pac. J. Trop. Med. 2011, 4, 589–593. [Google Scholar] [CrossRef] [Green Version]
- Elder, J.F.; Turner, B.J. Concerted Evolution of Repetitive DNA Sequences in Eukaryotes. Q. Rev. Biol. 1995, 70, 297–320. [Google Scholar] [CrossRef] [PubMed]
- De Gruijter, J.M.; Van Lieshout, L.; Gasser, R.B.; Verweij, J.J.; Brienen, E.A.; Ziem, J.B.; Yelifari, L.; Polderman, A.M. Polymerase chain reaction-based differential diagnosis of Ancylostoma duodenale and Necator americanus infections in humans in northern Ghana. Trop. Med. Int. Health 2005, 10, 574–580. [Google Scholar] [CrossRef]
- Verweij, J.J.; Pit, D.S.S.; Van Lieshout, L.; Baeta, S.M.; Dery, G.D.; Gasser, R.B.; Polderman, A.M. Determining the prevalence of Oesophagostomum bifurcum and Necator americanus infections using specific PCR amplification of DNA from faecal samples. Trop. Med. Int. Health 2001, 6, 726–731. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Geldhof, P.; Albonico, M.; Ame, S.M.; Bethony, J.M.; Engels, D.; Mekonnen, Z.; Montresor, A.; Sopheak, H.; Tchuem-Tchuenté, L.A. Molecular speciation of soil-transmitted helminths egg isolates collected during six drug efficacy trials in endemic countries. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 657–663. [Google Scholar] [CrossRef]
- Wang, A.G.; Dong, T.; Mansour, H.; Matamoros, G.; Sanchez, A.L.; Li, F. Paper-Based DNA Reader for Visualized Quantification of Soil-Transmitted Helminth Infections. ACS Sens. 2018, 3, 205–210. [Google Scholar] [CrossRef] [Green Version]
- Schär, F.; Odermatt, P.; Khieu, V.; Panning, M.; Duong, S.; Muth, S.; Marti, H.; Kramme, S. Evaluation of real-time PCR for Strongyloides stercoralis and hookworm as diagnostic tool in asymptomatic schoolchildren in Cambodia. Acta Trop. 2013, 126, 89–92. [Google Scholar] [CrossRef] [Green Version]
- Mejia, R.; Vicuña, Y.; Broncano, N.; Sandoval, C.; Vaca, M.; Chico, M.; Cooper, P.J.; Nutman, T.B. A Novel, Multi-Parallel, Real-Time Polymerase Chain Reaction Approach for Eight Gastrointestinal Parasites Provides Improved Diagnostic Capabilities to Resource-Limited At-Risk Populations. Am. J. Trop. Med. Hyg. 2013, 88, 1041–1047. [Google Scholar] [CrossRef]
- Basuni, M.; Muhi, J.; Othman, N.; Verweij, J.J.; Ahmad, M.; Miswan, N.; Rahumatullah, A.; Aziz, F.A.; Zainudin, N.S.; Noordin, R. A Pentaplex Real-Time Polymerase Chain Reaction Assay for Detection of Four Species of Soil-Transmitted Helminths. Am. J. Trop. Med. Hyg. 2011, 84, 338–343. [Google Scholar] [CrossRef] [Green Version]
- Llewellyn, S.; Inpankaew, T.; Nery, S.V.; Gray, D.J.; Verweij, J.J.; Clements, A.C.; Gomes, S.J.; Traub, R.; McCarthy, J.S. Application of a Multiplex Quantitative PCR to Assess Prevalence and Intensity Of Intestinal Parasite Infections in a Controlled Clinical Trial. PLoS Negl. Trop. Dis. 2016, 10, e0004380. [Google Scholar] [CrossRef]
- Taniuchi, M.; Verweij, J.J.; Noor, Z.; Sobuz, S.U.; Van Lieshout, L.; Petri, W.A., Jr.; Haque, R.; Houpt, E.R. High Throughput Multiplex PCR and Probe-based Detection with Luminex Beads for Seven Intestinal Parasites. Am. J. Trop. Med. Hyg. 2011, 84, 332–337. [Google Scholar] [CrossRef] [Green Version]
- Ngui, R.; Lim, Y.A.L.; Chua, K.H. Rapid Detection and Identification of Human Hookworm Infections through High Resolution Melting (HRM) Analysis. PLoS ONE 2012, 7, e41996. [Google Scholar] [CrossRef]
- Cunningham, L.J.; Stothard, J.R.; Osei-Atweneboana, M.; Armoo, S.; Verweij, J.J.; Adams, E.R. Developing a real-time PCR assay based on multiplex high-resolution melt-curve analysis: A pilot study in detection and discrimination of soil-transmitted helminth and schistosome species. Parasitology 2018, 145, 1733–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stracke, K.; Clarke, N.; Awburn, C.V.; Vaz Nery, S.; Khieu, V.; Traub, R.J.; Jex, A.R. Development and validation of a multiplexed-tandem qPCR tool for diagnostics of human soil-transmitted helminth infections. PLoS Negl. Trop. Dis. 2019, 13, e0007363. [Google Scholar] [CrossRef] [Green Version]
- Acosta Soto, L.; Santísima-Trinidad, A.B.; Bornay-Llinares, F.J.; Martín González, M.; Pascual Valero, J.A.; Ros Muñoz, M. Quantitative PCR and Digital PCR for Detection of Ascaris lumbricoides Eggs in Reclaimed Water. Biomed. Res. Int. 2017, 2017, 7515409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashwan, N.; Diawara, A.; Scott, M.E.; Prichard, R.K. Isothermal diagnostic assays for the detection of soil-transmitted helminths based on the SmartAmp2 method. Parasit. Vectors 2017, 10, 496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mugambi, R.M.; Agola, E.L.; Mwangi, I.N.; Kinyua, J.; Shiraho, E.A.; Mkoji, G.M. Development and evaluation of a Loop Mediated Isothermal Amplification (LAMP) technique for the detection of hookworm (Necator americanus) infection in fecal samples. Parasit. Vectors 2015, 8, 574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiraho, E.A.; Eric, A.L.; Mwangi, I.N.; Maina, G.M.; Kinuthia, J.M.; Mutuku, M.W.; Mugambi, R.M.; Mwandi, J.M.; Mkoji, G.M. Development of a Loop Mediated Isothermal Amplification for Diagnosis of Ascaris lumbricoides in Fecal Samples. J. Parasitol. Res. 2016, 2016, 7376207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papaiakovou, M.; Pilotte, N.; Baumer, B.; Grant, J.; Asbjornsdottir, K.; Schaer, F.; Hu, Y.; Aroian, R.; Walson, J.; Williams, S.A. A comparative analysis of preservation techniques for the optimal molecular detection of hookworm DNA in a human fecal specimen. PLoS Negl. Trop. Dis. 2018, 12, e0006130. [Google Scholar] [CrossRef]
- Ayana, M.; Cools, P.; Mekonnen, Z.; Biruksew, A.; Dana, D.; Rashwan, N.; Prichard, R.; Vlaminck, J.; Verweij, J.J.; Levecke, B. Comparison of four DNA extraction and three preservation protocols for the molecular detection and quantification of soil-transmitted helminths in stool. PLoS Negl. Trop. Dis. 2019, 13, e0007778. [Google Scholar] [CrossRef] [Green Version]
- Diawara, A.; Halpenny, C.M.; Churcher, T.S.; Mwandawiro, C.; Kihara, J.; Kaplan, R.M.; Streit, T.G.; Idaghdour, Y.; Scott, M.E.; Basáñez, M.G.; et al. Association between Response to Albendazole Treatment and β-Tubulin Genotype Frequencies in Soil-transmitted Helminths. PLoS Negl. Trop. Dis. 2013, 7, e2247. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, L.J.; Odoom, J.; Pratt, D.; Boatemaa, L.; Asante-Ntim, N.; Attiku, K.; Banahene, B.; Osei-Atweneboana, M.; Verweij, J.J.; Molyneux, D.; et al. Expanding molecular diagnostics of helminthiasis: Piloting use of the GPLN platform for surveillance of soil transmitted helminthiasis and schistosomiasis in Ghana. PLoS Negl. Trop. Dis. 2018, 12, e0006129. [Google Scholar] [CrossRef] [PubMed]
- Hii, S.F.; Senevirathna, D.; Llewellyn, S.; Inpankaew, T.; Odermatt, P.; Khieu, V.; Muth, S.; McCarthy, J.; Traub, R.J. Development and Evaluation of a Multiplex Quantitative Real-Time Polymerase Chain Reaction for Hookworm Species in Human Stool. Am. J. Trop. Med. Hyg. 2018, 99, 1186–1193. [Google Scholar] [CrossRef] [Green Version]
- Velasquez, D.E.; Arvelo, W.; Cama, V.A.; López, B.; Reyes, L.; Roellig, D.M.; Kahn, G.D.; Lindblade, K.A. Molecular Insights for Giardia, Cryptosporidium, and Soil-Transmitted Helminths from a Facility-Based Surveillance System in Guatemala. Am. J. Trop. Med. Hyg. 2011, 85, 1141–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunet, J.; Lemoine, J.P.; Lefebvre, N.; Denis, J.; Pfaff, A.W.; Abou-Bacar, A.; Traub, R.J.; Pesson, B.; Candolfi, E. Bloody Diarrhea Associated with Hookworm Infection in Traveler Returning to France from Myanmar. Emerg. Infect. Dis. 2015, 21, 1878–1879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng-Nguyen, D.; Hii, S.F.; Nguyen, V.-A.T.; Van Nguyen, T.; Van Nguyen, D.; Traub, R.J. Re-evaluation of the species of hookworms infecting dogs in Central Vietnam. Parasit. Vectors 2015, 8, 401. [Google Scholar] [CrossRef] [Green Version]
- Matamoros, G.; Rueda, M.M.; Rodríguez, C.; Gabrie, J.A.; Canales, M.; Fontecha, G.; Sanchez, A. High Endemicity of Soil-Transmitted Helminths in a Population Frequently Exposed to Albendazole but No Evidence of Antiparasitic Resistance. Trop. Med. Infect. Dis. 2019, 4, 73. [Google Scholar] [CrossRef] [Green Version]
- Roeber, F.; Jex, A.R.; Campbell, A.J.; Nielsen, R.; Anderson, G.A.; Stanley, K.K.; Gasser, R.B. Establishment of a robotic, high-throughput platform for the specific diagnosis of gastrointestinal nematode infections in sheep. Int. J. Parasitol. 2012, 42, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Sanprasert, V.; Kerdkaew, R.; Srirungruang, S.; Charuchaibovorn, S.; Phadungsaksawasdi, K.; Nuchprayoon, S. Development of Conventional Multiplex PCR: A Rapid Technique for Simultaneous Detection of Soil-Transmitted Helminths. Pathogens 2019, 8, 152. [Google Scholar] [CrossRef] [Green Version]
- Areekul, P.; Putaporntip, C.; Pattanawong, U.; Sitthicharoenchai, P.; Jongwutiwes, S. Trichuris vulpis and T. trichiura infections among schoolchildren of a rural community in northwestern Thailand: The possible role of dogs in disease transmission. Asian Biomed. 2010, 4, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Phuphisut, O.; Yoonuan, T.; Sanguankiat, S.; Chaisiri, K.; Maipanich, W.; Pubampen, S.; Komalamisra, C.; Adisakwattana, P. Triplex polymerase chain reaction assay for detection of major soil-transmitted helminths, Ascaris lumbricoides, Trichuris trichiura, Necator americanus, in fecal samples. Southeast Asian J. Trop. Med. Public Health 2014, 45, 267–275. [Google Scholar]
- Pilotte, N.; Maasch, J.R.M.A.; Easton, A.V.; Dahlstrom, E.; Nutman, T.B.; Williams, S.A. Targeting a highly repeated germline DNA sequence for improved real-time PCR-based detection of Ascaris infection in human stool. PLoS Negl. Trop. Dis. 2019, 22, e0007593. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-X.; Pan, C.-S.; Cui, L.-W. Application of a real-time PCR method for detecting and monitoring hookworm Necator americanus infections in Southern China. Asian Pac. J. Trop. Biomed. 2012, 2, 925–929. [Google Scholar] [CrossRef] [Green Version]
- Keller, L.; Patel, C.; Welsche, S.; Schindler, T.; Hürlimann, E.; Keiser, J. Performance of the Kato-Katz method and real time polymerase chain reaction for the diagnosis of soil-transmitted helminthiasis in the framework of a randomised controlled trial: Treatment efficacy and day-to-day variation. Parasit. Vectors 2020, 13, 1–12. [Google Scholar] [CrossRef]
- Dunn, J.C.; Papaiakovou, M.; Han, K.T.; Chooneea, D.; Bettis, A.A.; Wyine, N.Y.; Lwin, A.M.M.; Maung, N.S.; Misra, R.; Littlewood, D.T.J.; et al. The increased sensitivity of qPCR in comparison to Kato-Katz is required for the accurate assessment of the prevalence of soil-transmitted helminth infection in settings that have received multiple rounds of mass drug administration. Parasit Vectors. 2020, 13, 324. [Google Scholar] [CrossRef] [PubMed]
- Barda, B.; Schindler, C.; Wampfler, R.; Ame, S.; Ali, S.M.; Keiser, J. Comparison of real-time PCR and the Kato-Katz method for the diagnosis of soil-transmitted helminthiasis and assessment of cure in a randomized controlled trial. BMC Microbiol. 2020, 20, 298. [Google Scholar] [CrossRef] [PubMed]
- Levecke, B.; Cools, P.; Albonico, M.; Ame, S.; Angebault, C.; Ayana, M.; Behnke, J.M.; Bethony, J.M.; Cringoli, G.; Dana, D.; et al. Identifying thresholds for classifying moderate-to-heavy soil-transmitted helminth intensity infections for FECPAKG2, McMaster, Mini-FLOTAC and qPCR. PLoS Negl. Trop. Dis. 2020, 14, e0008296. [Google Scholar] [CrossRef]
- Cools, P.; Van Lieshout, L.; Koelewijn, R.; Addiss, D.; Ajjampur, S.S.; Ayana, M.; Bradbury, R.S.; Cantera, J.L.; Dana, D.; Fischer, K.; et al. First international external quality assessment scheme of nucleic acid amplification tests for the detection of Schistosoma and soil-transmitted helminths, including Strongyloides: A pilot study. PLoS Negl. Trop. Dis. 2020, 14, e0008231. [Google Scholar] [CrossRef]
- Cools, P.; Vlaminck, J.; Verweij, J.J.; Levecke, B. Quantitative PCR in soil-transmitted helminth epidemiology and control programs: Toward a universal standard. PLoS Negl. Trop. Dis. 2021, 15, e0009134. [Google Scholar] [CrossRef]
- Papaiakovou, M.; Gasser, R.B.; Littlewood, D.T.J. Quantitative PCR-Based Diagnosis of Soil-Transmitted Helminth Infections: Faecal or Fickle? Trends Parasitol. 2019, 35, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Papaiakovou, M.; Wright, J.; Pilotte, N.; Chooneea, D.; Schär, F.; Truscott, J.E.; Dunn, J.C.; Gardiner, I.; Walson, J.L.; Williams, S.A.; et al. Pooling as a strategy for the timely diagnosis of soil-transmitted helminths in stool: Value and reproducibility. Parasit. Vectors. 2019, 12, 443. [Google Scholar] [CrossRef] [Green Version]
- Truscott, J.E.; Dunn, J.C.; Papaiakovou, M.; Schaer, F.; Werkman, M.; Littlewood, D.T.J.; Walson, J.L.; Anderson, R.M. Calculating the prevalence of soil-transmitted helminth infection through pooling of stool samples: Choosing and optimizing the pooling strategy. PLoS Negl. Trop. Dis. 2019, 13, e0007196. [Google Scholar] [CrossRef]
- Leta, G.T.; French, M.; Dorny, P.; Vercruysse, J.; Levecke, B. Comparison of individual and pooled diagnostic examination strategies during the national mapping of soil-transmitted helminths and Schistosoma mansoni in Ethiopia. PLoS Negl. Trop. Dis. 2018, 12, e0006723. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashwan, N.; Bourguinat, C.; Keller, K.; Gunawardena, N.K.; De Silva, N.; Prichard, R. Isothermal Diagnostic Assays for Monitoring Single Nucleotide Polymorphisms in Necator americanus Associated with Benzimidazole Drug Resistance. PLoS Negl. Trop. Dis. 2016, 10, e0005113. [Google Scholar] [CrossRef]
- Pomari, E.; Piubelli, C.; Perandin, F.; Bisoffi, Z. Digital PCR: A new technology for diagnosis of parasitic infections. Clin. Microbiol. Infect. 2019, 25, 1510–1516. [Google Scholar] [CrossRef] [PubMed]
- Weerakoon, K.G.; McManus, D.P. Cell-Free DNA as a Diagnostic Tool for Human Parasitic Infections. Trends Parasitol. 2016, 32, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Gorgani-Firouzjaee, T.; Kalantari, N.; Javanian, M.; Ghaffari, S. Strongyloides stercoralis: Detection of parasite-derived DNA in serum samples obtained from immunosuppressed patients. Parasitol. Res. 2018, 117, 2927–2932. [Google Scholar] [CrossRef] [PubMed]
- Lodh, N.; Caro, R.; Sofer, S.; Scott, A.; Krolewiecki, A.; Shiff, C. Diagnosis of Strongyloides stercoralis: Detection of parasite-derived DNA in urine. Acta Trop. 2016, 163, 9–13. [Google Scholar] [CrossRef] [PubMed]
| Method | Principle | Sensitivity | Negative Predictive Value | Reference |
|---|---|---|---|---|
| Wet mount preparation | Preparation of stool sample with saline/iodine on a microscopic slide with a cover glass. | Hookworm: 85.7% A. lumbricoides: 83.3% | Hookworm: 97.5% A. lumbricoides: 98.8% | [8] |
| Hookworm: 37.9% A. lumbricoides: 52% T. trichiura:1 2.5% | [9] | |||
| Formol–ether sedimentation | Stool sample diluted in distilled water is centrifuged with 3% ethyl ether. Of the four layers formed, the lower sediment is mixed with 5% formalin and 50 uL of this sediment screened under a microscope. | Hookworm: 95.8% A. lumbricoides: 94.2% T. trichiura: 86.7% | Hookworm: 98.8% A. lumbricoides: 98.5% T. trichiura: 99.8% | [21] |
| Hookworm: 72.4% A. lumbricoides: 81.4% T. trichiura: 57.8% | [9] | |||
| Kato–Katz | A known amount of sieved stool sample is placed on a glass slide and covered with cellophane soaked in methylene blue solution and screened under a microscope after 30 min and the eggs are counted. | Hookworm: 69% A. lumbricoides: 93.1% T. trichiura: 90.6% | [9] | |
| Hookworm: 72.4% A. lumbricoides: 57.4% Trichuris spp.: 84.4% | [10] | |||
| Hookworm: 62.9% A. lumbricoides: 72.4% T. trichiura: 95% | Hookworm: 58.8% A. lumbricoides: 81.3% T. trichiura: 50% | [22] | ||
| Hookworm: 81% A. lumbricoides: 68% T. trichiura: 88% | [15] | |||
| A. lumbricoides: 84.4% | A. lumbricoides: 96.7% | [23] | ||
| Hookworm: 89.1% A. lumbricoides: 89.8% T. trichiura: 96.1% | [17] | |||
| Hookworm: 19.6% A. lumbricoides: 67.8% T. trichiura: 76.6% | Hookworm: 84.3% A. lumbricoides: 86.7% T. trichiura: 75.3% | [14] | ||
| Hookworm: 93.9% A. lumbricoides: 96.2% T. trichiura: 98.3% | [24] | |||
| Hookworm: 95.1% A. lumbricoides: 97.3% T. trichiura: 86.7% | Hookworm: 98.6% A. lumbricoides: 99.3% T. trichiura: 99.8% | [21] | ||
| McMaster | A known amount of stool is mixed with a saturated salt solution. The top layer of the solution is added to the reading chamber and the eggs are counted. | Hookworm: 67.6% A. lumbricoides: 74.3% T. trichiura: 94.6% | Hookworm: 62% A. lumbricoides: 82.4% T. trichiura: 47.8% | [22] |
| Hookworm: 92.3% A. lumbricoides: 94.9% T. trichiura: 95.2% | [24] | |||
| A. lumbricoides: 48.3% | A. lumbricoides: 91.5% | [23] | ||
| Hookworm: 80.8% A. lumbricoides: 69.5% T. trichiura: 46.7% | Hookworm: 94.6% A. lumbricoides: 92.7% T. trichiura: 99.4% | [21] | ||
| FLOTAC | A weighed amount of stool sample is homogenized and filtered with water or saline. The filtrate is mixed with a floatation solution and is added to the two FLOTAC chambers. The chambers are centrifuged and examined under a microscope. | Hookworm: 80.8% A. lumbricoides: 81.9% T. trichiura: 96.8% | Hookworm: 73.4% A. lumbricoides: 86.9% T. trichiura: 61.1% | [22] |
| Hookworm: 54% A. lumbricoides: 88% T. trichiura: 95% | [15] | |||
| Hookworm: 100% A. lumbricoides: 100% T. trichiura: 100% | Hookworm: 100% A. lumbricoides: 100% T.t richiura: 100% | [14] | ||
| Mini-FLOTAC | A weighed stool sample is homogenized with 5% formalin, filtered, and the filtrate is added to the flotation solution. The suspension is loaded into the chamber. | A. lumbricoides: 61.3% | A. lumbricoides: 93.1% | [23] |
| Hookworm: 70.8% A. lumbricoides: 42.1% T. trichiura: 85.6% | [10] | |||
| FECPAKG2 | FECPAKG2 platform with a cassette, concentrates helminth eggs into one microscopic field of view and this is photographed and is stored on a computer from which the eggs can be counted. | Hookworm: 47.5% A. lumbricoides: 36.8% T. trichiura: 37.5% | [10] | |
| Hookworm: 91.3% A. lumbricoides: 96.9% T. trichiura: 95.3% | [17] |
| Molecular Technique | Target | STH Detected | Sensitivity * | Negative Predictive Value * | Reference | |
|---|---|---|---|---|---|---|
| Conventional PCR | Single-plex | Mitochondrial COI gene | A. duodenale, N. americanus | [36] | ||
| Nested PCR | Semi-nested PCR | ITS-2 | A. duodenale | [40] | ||
| 28S rRNA, ITS-2 | N. americanus | N. americanus: 94.5% | [41] | |||
| Semi-nested PCR+ RFLP | ITS-1, 2 and 5.8S region | A. duodenale, N. americanus, | [42] | |||
| T. trichiura, A. lumbricoides | ||||||
| Quantitative paper-based DNA reader | Single-plex mini-PCR | β-tubulin | T. trichiura | [43] | ||
| Real-time PCR | Single-plex | ITS-2 | A. duodenale, N. americanus | Hookworm: 78.9% | [44] | |
| Repetitive sequence | N. americanus, A. lumbricoides, T. trichiura, A. duodenale | [35] | ||||
| Multiparallel | ITS-1,2 | A. lumbricoides, T. trichiura, A. duodenale, N. americanus | A. lumbricoides: 96.7% T. trichiura: 99.2% | [45] | ||
| Repetitive sequences, ITS-1,2 | A. lumbricoides, T. trichiura, A. duodenale, N. americanus | A. lumbricoides: 98% N. americanus: 98% | [28] | |||
| Multiplex | ITS-2 | A. duodenale, N. americanus | [34] | |||
| ITS-1,2 | A. lumbricoides, Ancylostoma spp., N. americanus | [46] | ||||
| ITS-1,2 | N. americanus, T. trichiura, Ancylostoma spp., Ascaris spp. | [47] | ||||
| PCR-Luminex | ITS-1,2 | N. americanus, A. lumbricoides, A. duodenale | [48] | |||
| Melt curve analysis | ITS-2 | N. americanus, A. duodenale, A. ceylanicum, A. caninum, A. braziliense | Hookworm: 100% | [49] | ||
| 18S, ITS-1,2 | A. lumbricoides, T. trichiura, A. duodenale, N. americanus | [50] | ||||
| Multiplex-tandem PCR–qPCR | β-tubulin | A. lumbricoides, T. trichiura, A. duodenale, N. americanus | [51] | |||
| Digital PCR | ITS-1 | A. lumbricoides | [52] | |||
| Isothermal assay | SmartAmp2 | β-tubulin | N. americanus, A. lumbricoides, T. trichiura | [53] | ||
| LAMP | ITS-2 | N. americanus | N. americanus: 97% | [54] | ||
| LAMP | ITS-1 | A. lumbricoides | A. lumbricoides: 96.3% | A. lumbricoides: 88.9% | [55] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manuel, M.; Ramanujam, K.; Ajjampur, S.S.R. Molecular Tools for Diagnosis and Surveillance of Soil-Transmitted Helminths in Endemic Areas. Parasitologia 2021, 1, 105-118. https://doi.org/10.3390/parasitologia1030012
Manuel M, Ramanujam K, Ajjampur SSR. Molecular Tools for Diagnosis and Surveillance of Soil-Transmitted Helminths in Endemic Areas. Parasitologia. 2021; 1(3):105-118. https://doi.org/10.3390/parasitologia1030012
Chicago/Turabian StyleManuel, Malathi, Karthik Ramanujam, and Sitara S. R. Ajjampur. 2021. "Molecular Tools for Diagnosis and Surveillance of Soil-Transmitted Helminths in Endemic Areas" Parasitologia 1, no. 3: 105-118. https://doi.org/10.3390/parasitologia1030012
APA StyleManuel, M., Ramanujam, K., & Ajjampur, S. S. R. (2021). Molecular Tools for Diagnosis and Surveillance of Soil-Transmitted Helminths in Endemic Areas. Parasitologia, 1(3), 105-118. https://doi.org/10.3390/parasitologia1030012