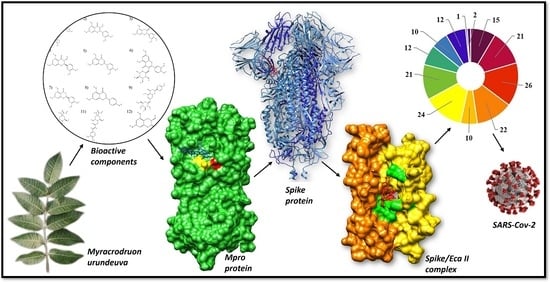
Bioactive Components of Myracrodruon urundeuva against SARS-CoV-2: A Computational Study
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Selection of Chemical Compounds of Myracrodruon urundeuva
3.2. Determination of the Active Site
3.3. Molecular Docking Study
3.4. ADME-TOX Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dourado, L.; Caetano, L.; Marques, M.; Penna, U.; Costa, C.; Arruda, F.; Libera, L. Estudo da história natural da COVID-19 e epidemiology of SARS-CoV-2 infection: A descriptive review of the literature. Braz. J. Surg. Clin. Res. 2020, 33, 46–56. [Google Scholar]
- Oliveira, M.; Robinson, A.; Siqueira, M. Knowing SARS-CoV-2 and COVID-19. In Health Diplomacy and COVID-19: Reflections Halfway, 1st ed.; Fonseca, L., Ed.; Fiocruz: São Paulo, Brazil, 2020; pp. 69–82. [Google Scholar]
- Johns Hopkins Coronavirus Resource Center. Available online: https://coronavirus.jhu.edu/map.html (accessed on 11 November 2022).
- Nascimento, L.; Marchiori, M.; Field, V.; Zini, M. SARS-CoV-2 and COVID-19: Pathophysiological and immunological aspects, diagnostic strategies and vaccine development. Rev. Interdiscip. Saúde Educ. 2020, 1, 122–158. [Google Scholar] [CrossRef]
- Schoeman, D.; Fielding, B. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef]
- Lin, L.; Lu, L.; Cao, W.; Li, T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 2020, 9, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Marra, M.A.; Jones, S.J.M.; Astell, C.R.; Holt, R.A.; Brooks-Wilsin, A.; Butterfield, Y.S.N.; Khattra, J.; Asano, J.K.; Barberb, S.A.; Chan, S.Y.; et al. The Genome Sequence of the SARS-Associated Coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, I.; Galvão, M.; Souza, T.; Gomes, S.; Medeiros, A.; Lima, K. Incidence of and mortality from COVID-19 in the older Brazilian population and its relationship with contextual indicators: An ecological study. Rev. Bras. Geriatr. Gerontol. 2020, 23, 2017. [Google Scholar] [CrossRef]
- Antonio, A.; Wiedemann, L.; Junior, V. Natural products’ role against COVID-19. RSC Adv. 2020, 10, 23379–23393. [Google Scholar] [CrossRef]
- Domingos, F.; Silva, M. Use, knowledge and conservation of Myracrodruon urundeuva: A systematic review. Res. Soc. Dev. 2020, 9, 8851. [Google Scholar] [CrossRef]
- Amaral, E.A.; Silva, R.M.G. Evaluation of the acute toxicity of angico (Anadenanthera falcata), pau-santo (Kilmeyera coreacea), aroeira (Myracrodruon urundeuva) and São João vine (Pyrostegia venusta), by means of the bioassay with Artemia salina. J. Serv. Res. 2008, 5, 1–16. [Google Scholar]
- Maia, G. Caatinga, Trees and Shrubs and Their Utilities, 1st ed.; D & Z: São Paulo, Brazil, 2004. [Google Scholar]
- Souza, S.; Aquino, L.; Milach, A.; Bandeira, M.; Nobre, M.; Viana, G. Antiinflammatory and antiulcer properties of tannins from Myracrodruon urundeuva Allemão (Anacardiaceae) in Rodents. Phytother. Res. 2007, 21, 2020–2225. [Google Scholar] [CrossRef]
- Asadi, S.; Ahmadiani, A.; Esmaeili, M.; Sonboli, A.; Ansari, N.; Khodagholi, F. In vitro antioxidant activities and an investigation of neuroprotection by six Salvia species from Iran: A comparative study. FTC 2010, 48, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Filho, F.; Nascimento, K.; Santos, W.; Frazão, N. Study of acetylcholinesterase inhibition by molecular docking: Application in the treatment of Alzheimer’s disease. Ed. S. Health. 2020, 7, 1–18. [Google Scholar] [CrossRef]
- Ferreira, L.; Santos, R.; Oliva, G.; Andricopulo, A. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Bankaitis, V. Molecular Docking: From Lock and Key to Combination Lock. J. Mol. Med. Clin. Appl. 2017, 2, 106. [Google Scholar] [CrossRef]
- Dias, R.; Azevedo, J.; Walter, F. Molecular Docking Algorithms. Curr. Drug Targets 2008, 9, 1040–1047. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.B. Chemical Profile and Cytotoxic Activity of Leaf, Branch and Bark Extracts of Aroeira-Do-Sertão (Myracrodruon urundeuva All.): Metabolomic and Chemometric Approach; Monograph (Graduation in Chemistry); Federal University of Ceará: Fortaleza, Brazil, 2016. [Google Scholar]
- Figueredo, F.; Lucena, B.; Tintino, S.; Matias, E.; Leite, N.; Andrade, J.; Nogueira, L.; Morais, E.; Costa, J.; Coutinho, H.; et al. Chemical composition and evaluation of modulatory of the antibiotic activity from extract and essential oil of Myracrodruon urundeuva. Pharm. Biol. 2014, 52, 560–565. [Google Scholar] [CrossRef]
- Costa, O.B.; Mennezi, C.; Benedito, L.; Resck, I.; Vieria, F.; Bizzo, H. Essential oil constituents and yields from leaves of Blepharocalyx salicifolius (Kunt) O. Berg and Myracrodruon urundeuva (Allemão) collected during daytime. J. For. Res. 2014, 2014, 982576. [Google Scholar] [CrossRef]
- Alquino, N. Chemical Aspects of the Chemical-Pharmacological-Agronomic Study of Wild and Cultivated Aroeiras do Sertão (Myracrodruon urundeuva Fr. all). Ph.D. Thesis, Federal University of Ceará, Fortaleza, Brazil, 2017. [Google Scholar]
- Pessoa, E.; Nicolete, R.; Araújo, T. The use of Anacardiaceae Family’s vegetable extracts as natural skin-clearing agents: A review. World J. Pharm. Res. 2020, 9, 32–37. [Google Scholar] [CrossRef]
- Ruyck, J.; Brysbaert, G.; Blossey, R.; Lensink, M. Molecular docking as a popular tool in drug design, an in silico travel. Adv. Appl. Bioinform. Chem. 2016, 9, 1–11. [Google Scholar] [CrossRef]
- Smith, R.; Cavalcante, G. Potential therapeutic targets for COVID-19: An integrative review. JCS HU-UFPI 2021, 4, 18–25. [Google Scholar] [CrossRef]
- Oliveira, Y.R.; Robinson, W.; Smith, P.; Pacheco, A.; Abreu, M. Anacardiaceae in the Traditional Medicine of Rural Communities of Piauí, Northeastern Brazil. Ens. Ciências 2022, 26, 32–42. [Google Scholar] [CrossRef]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.; Hjorth, C.; Dommer, A.; Harbison, A.; Fogarty, C.; Barros, E.; Taylor, B.; McLellan, J.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS. Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef] [PubMed]
- Junior, E.; Gonsalves, A. Molecular docking study of gsk-3β enzyme inhibitors: A proposal for the treatment of bipolar disorder. Rev. Ifes Sci. 2019, 5, 243–256. [Google Scholar] [CrossRef]
- Ord, M.; Faustova, I.; Loog, M. The sequence at Spike S1/S2 site enables cleavage by furin and phospho-regulation in SARS-CoV-2 but not in SARS-CoV-1 or MERS-CoV. Sci. Rep. 2020, 10, 16944. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Li, Y.; Zeng, R.; Liu, F.; Luo, R.; Huang, C.; Wang, Y.; Zhang, J.; Quan, B.; Yang, S. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science 2019, 371, 1374–1378. [Google Scholar] [CrossRef]
- Kumar, S. COVID-19: A Drug Repurposing and Biomarker Identification by Using Comprehensive Gene-Disease Associations through Protein-Protein Interaction Network Analysis. Preprints 2020, 1, 2020030440. [Google Scholar] [CrossRef]
- Bissantz, C.; Kuhn, B.; Stahl, M. A medicinal chemist’s guide to molecular interactions. J. Med. Chem. 2010, 22, 5061–5084. [Google Scholar] [CrossRef]
- Incerti, M.K.; Rosa, T.F.; Foletto, V.S.; Franco, L.N.; Hörner, R. Repurposing Medications for COVID-19: An Overview. Health 2022, 48, 1–5. [Google Scholar]
- Robinson, M.; Facchainetti, V.; Aboud, K.; Penha, L.; Gomes, C. Repositioning of the drug baricitinib for the treatment of COVID-19. Conjectures 2022, 22, 306–321. [Google Scholar] [CrossRef]
- Smith, D.; Padilha, I. In silico evaluation of pharmacokinetic properties of antileukemic compounds published by scientific journals. Arch. Health. Sci. 2022, 30, e04978. [Google Scholar] [CrossRef]
- Lennernäs, H.; Abrahamsson, B. The use of biopharmaceutic classification of drugs in drug discovery and development: Current status and future extension. J. Pharm. Pharmacol. 2005, 57, 273–410. [Google Scholar] [CrossRef]
- Jorgensen, W.; Duffy, E. Prediction of drug solubility from structure. Adv. Drug Deliv. Rev. 2002, 54, 355–366. [Google Scholar] [CrossRef]
- Robinson, J.; Robinson, Z.; Storpirtis, S. In vitro models for determining drug absorption and predicting the dissolution/absorption ratio. Rev. Bras. Cienc. Farm. 2007, 43, 515–527. [Google Scholar] [CrossRef]
- Fernandes, M.B.; Gonçalves, J.; Scotti, M.; Oliveira, A.; Tavares, L.; Storpirtis, S. Caco-2 cells cytotoxicity of nifuroxazide derivatives with potential activity against Methicillin-resistant Staphylococcus aureus (MRSA). Toxicol. In Vitr. 2012, 26, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Meanwell, N.A. Improving Drug Candidates by Design: A Focus on Physicochemical Properties as a Means of Improving Compound Disposition and Safety. Chem. Res. Toxicol. 2011, 24, 1420–1456. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, R.; Júnior, A.; Zepeda, C.; Santos, L.; Pinto, L.; Cabral, O.; Soto, C. In silico analysis of the pharmacokinetic and toxicological profile of Zinc II thioglycolate complex [Zn(ATG)2(OH2)2]. Res. Soc. Dev. 2022, 11, e44711629430. [Google Scholar] [CrossRef]
- Holguín, N.M.; Frau, J.; Mitnik, D.G. Computational Pharmacokinetics Report, ADMET Study and Conceptual DFT-Based Estimation of the Chemical Reactivity Properties of Marine Cyclopeptides. Chem. Open 2021, 10, 1087–1157. [Google Scholar] [CrossRef]
- Aiub, C.A.F.; Felzenszwalb, I. The principles of the ames (Salmonella/microsome) test and its applicability. Genet. Sch. 2011, 6, 11–16. [Google Scholar] [CrossRef]
- Pimentel, L.C.F.; Keys, C.; Robinson, L.; Alfonso, J. The unbelievable use of dangerous chemicals in the past. Whoops. New. 2006, 29, 1138–1149. [Google Scholar] [CrossRef]
- Bertolami, M.C. Mechanisms of hepatotoxicity. Arq. Bras. Cardiol. 2005, 85, 25–27. [Google Scholar] [CrossRef]
- Li, Y.; Chi, W.; Su, J.; Ferrall, L.; Hung, C.; Wu, T. Coronavirus vaccine development: From SARS and MERS to COVID-19. J. Biomed. Sci. 2020, 27, 104. [Google Scholar] [CrossRef]
- Tao, K.; Tzou, F.L.; Nouhin, J.; Gupta, R.K.; Oliveira, T.; Kosakovsky, L.S.L.; Fera, D.; Shafer, R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef]
- World Health Organization. Available online: https://www.who.int/docs/default-source/coronaviruse/09082023eg.5_ire_final.pdf (accessed on 4 September 2023).
- Robinson, R.; Fagundes, M.; Roberts, H.; Veloso, M. Ecological aspects of aroeira (Myracrodruon urundeuva Allemão- Anacardiaceae): Phenology and seed germination. Rev. Árvore 2008, 32, 233–243. [Google Scholar] [CrossRef]
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 10 August 2022).
- Morais, J.P.A.; Pappa, G.L.; Pires, D.E.V.; Izidoro, S.C. GASS-WEB: A web server for identifying enzyme active sites based on genetic algorithms. Nucleic Acids Res. 2017, 45, 315–319. [Google Scholar] [CrossRef]
- Moraes, J.P.A.; Izidoro, S.C.; Melo-Minardi, R.C.; Pappa, G.L. GASS: Identifying enzyme active sites with genetic algorithms. Bioinformatics 2015, 31, 864–870. [Google Scholar] [CrossRef]
- RCSB Protein Data Bank. Available online: http://www.rcsb.org/ (accessed on 16 September 2022).
- Barros, R.O.; Junior, F.; Perreira, W.; Oliveira, N.; Ramos, R. Interaction of Drug Candidates with Various SARS-CoV-2 Receptors: An in-Silico Study to Combat COVID-19. J. Proteome Res. 2020, 19, 4567–4575. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Golddard, T.; Huang, C.; Couch, G.; Greenblatt, D.; Meng, L.; Ferrin, T. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- pkCSM: Predicting Small-Molecule Pharmacokinetic Properties Using Graph-Based Signatures. Available online: https://biosig.lab.uq.edu.au/pkcsm/ (accessed on 12 February 2023).
- Rocha, J.A.; Rego, N.; Carvalho, B.; Silva, F.; Sousa, J.; Ramos, R.; Passos, I.; Maraes, J.; Lima, F. Computational quantum chemistry, molecular docking, and ADMET predictions of imidazole alkaloids of Pilocarpus microphyllus with schistosomicidal properties. PLoS ONE 2018, 13, e0198476. [Google Scholar] [CrossRef] [PubMed]







| Complex (Ligand–Protein) | ΔGbinda (kcal.mol−1) | Amino Acids That Interact by Hydrogen Bonding | Amino Acids That Interact by Hydrophobic Bonding |
|---|---|---|---|
| Agathisflavone/Spike | −9.7 | His519, His49, Ser967, Asp571 | Val42, Asp40, Asp568, Agr567, Agr44, Lys964, Leus518 |
| Agathisflavone/RBD | −9.7 | Lys417, Asn33, Asp30, Phe390, Ser494, Asp405 | Ala387, Pre389, Leu455, Tyr495, Agr403, Asp38, Tyr453, His34, Glu37, Arg393, Tyr505, Ala386 |
| Agathisflavone/MPro | −9.3 | Glu166, Phe140 | Thr190, Gln189, His41, Met49, Met165, His164, Cys145, Leu141 |
| Quercetin/Spike | −9.0 | Arg100, Leu977, Thr573, Phe855, Tyr741 | Gly744, Leu966, Val976, Thr547, Leu546, Asn978, Thr572, Met740, Asn856 |
| Gallocatechin gallate /MPro | −9.0 | Phe140, Thr26, Ser144, His163, Leu141, Glu166 | His172, Asn142, Leu27, Gly143, Thr25, Cys145, Met49, His41, Arg188, Asp187, Gln189, Met165 |
| Taxifolin/Spike | −9.0 | Ile742, Tyr741, Asn978 | Ile587, Thr573, Val976, Leu977, Gly744, Arg1000, Leu966, Thr572, |
| Luteolin/Spike | −9.0 | Met740, Phe855, Thr573, Arg1000, Tyr741 | Gly744, Asn856, Gly548, Asn978, Thr547, Leu546, Val976, Thr527, Leu966 |
| Quercitrin/MPro | −9.0 | Asp187, Asn142, Leu141, Ser144, His163, Glu166, Tyr54 | Arg188, Met49, His164, Met165, Gly143, Cys145, Gln168, His41 |
| Eriodictiol/Spike | −8.9 | Asn856, Ile742, Tyr741 | Ile587, Asn978, Val976, Leu977, Gly744, Arg1000, Thr572, Thr573 |
| Apigenin/Spike | −8.7 | Phe855, Thr573, Arg1000, Tyr741, Met740, | Asn856, Gly744, Asn978, Leu546, Thr547, Val976, Thr572, Leu966 |
| Gallocatechin gallate/Spike | −8.7 | His49, Arg44, Asp40, Asp571, Ser968, Ser967, | Asp568, Ile569, Arg567, Gly757, Asn969, Leu754, Gln755 |
| Naringenin/Spike | −8.7 | Val976, Ser974, Arg983, Ile973, Asp979, Ser975 | His519, Arg567, Thr430, Leu518, Glu516, Asp571, Asn969, |
| Cryptochlorogenic acid/Spike | −8.6 | Arg1000, Ser975, Leu977, Asn978, Asn856, Ala570, Leu966 | Val976, Pro589, Thr573, Phe855, Thr572 |
| Feruloyl-D-quinic acid/Spike | −8.6 | Arg1000, Ser975, Leu977, Asn978, Asp568, Thr549, Gly744 | Asn856, Thr572, Ile587, Phe589, Phe855, Gly548, Thr573 |
| Gallocatechin/Spike | −8.6 | Phe855, Thr549, Thr573, Thr572, Leu977, Gly744, Arg1000, Tyr741, Met740, | Pre589, Ile587, Asn856, Phe541 |
| Compounds | ΔGbinda (kcal.mol−1) | |||
|---|---|---|---|---|
| ACE2 Protein | Mpro Protein | RBD Protein | Spike Protein | |
| Baricitinib | −6.8 | −7.9 | −7.8 | −8.0 |
| Molnupiravir | −7.2 | −6.7 | −6.8 | −7.9 |
| Remdesivir | −7.3 | −7.9 | −7.6 | −7.5 |
| Compounds | Solubility in Water (log mol.L−1) | PCaco2 (Log Papp in 10−6 cm.s−1) | AIH% | P.Skin (log Kp) |
|---|---|---|---|---|
| Agathisflavone | −2.892 | 0.371 | 94.062 | −2.735 |
| Apigenin | −3.178 | 1.076 | 91.856 | −2.736 |
| Cryptochlorogenic acid | −2.854 | −0.707 | 15.087 | −2.735 |
| Eriodictiol | −3.344 | 0.787 | 79.846 | −2.736 |
| Feruloyl-D-quinic acid | −2.776 | −0.576 | 19.764 | −2.735 |
| Gallocatechin gallate | −2.895 | −0.797 | 57.176 | −2.735 |
| Luteolin | −3.173 | 0.762 | 81.082 | −2.735 |
| Naringenin | −3.903 | 0.634 | 68.462 | −2.735 |
| Quercetin | −2.982 | 0.694 | 74.84 | −2.735 |
| Quercitrin | −3.132 | −0.476 | 55.404 | −2.735 |
| Taxifolin | −3.031 | −0.318 | 70.529 | −2.735 |
| Compounds | VDss (Human) (log L.Kg−1) | P.B.H (log BB) |
|---|---|---|
| Agathisflavone | −0.943 | −2.192 |
| Apigenin | −0.105 | −0.951 |
| Cryptochlorogenic acid | −1.495 | −1.737 |
| Eriodictiol | 0.229 | −1.180 |
| Feruloyl-D-quinic acid | −1.738 | −1.593 |
| Gallocatechin gallate | 0.050 | −2.209 |
| Luteolin | 0.071 | −1.199 |
| Naringenin | −0.431 | −1.449 |
| Quercetin | 0.310 | −1.377 |
| Quercitrin | −0.315 | −2.027 |
| Taxifolin | 0.547 | −1.328 |
| Compounds | T.AMES | D.M.T (log mg.kg−1.day−1) | T.A.O (LD50) (mol.kg−1) | T.C.O (LOAEL) (log mg.kg−1.day−1) | S.Skin | Hep |
|---|---|---|---|---|---|---|
| Agathisflavone | No | 0.425 | 2.467 | 3.285 | No | No |
| Apigenin | No | 0.931 | 2.376 | 1.461 | No | No |
| Cryptochlorogenic acid | No | 1.379 | 2.219 | 3.503 | No | No |
| Eriodictiol | Yes | 0.395 | 2.229 | 1.893 | No | No |
| Feruloyl-D-quinic acid | No | 1.428 | 2.133 | 3.587 | No | No |
| Gallocatechin gallate | Yes | 0.481 | 2.654 | 4.085 | No | No |
| Luteolin | No | 0.975 | 2.450 | 1.833 | No | No |
| Naringenin | Yes | 0.989 | 3.573 | 3.556 | No | No |
| Quercetin | Yes | 0.954 | 2.308 | 3.134 | No | No |
| Quercitrin | Yes | 0.878 | 2.930 | 2.826 | No | No |
| Taxifolin | Yes | 0.886 | 2.245 | 3.256 | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, S.K.S.; Sousa, C.S.; Viana, E.K.A.; Souza, H.C.A.; Souza, M.D.A.; Ribeiro, A.S.N.; Vale, V.d.S.d.; Islam, M.T.; Araújo, J.L.; Rocha, J.A. Bioactive Components of Myracrodruon urundeuva against SARS-CoV-2: A Computational Study. Drugs Drug Candidates 2023, 2, 781-795. https://doi.org/10.3390/ddc2040039
Alves SKS, Sousa CS, Viana EKA, Souza HCA, Souza MDA, Ribeiro ASN, Vale VdSd, Islam MT, Araújo JL, Rocha JA. Bioactive Components of Myracrodruon urundeuva against SARS-CoV-2: A Computational Study. Drugs and Drug Candidates. 2023; 2(4):781-795. https://doi.org/10.3390/ddc2040039
Chicago/Turabian StyleAlves, Sabrina Kelly Silva, Cássio Silva Sousa, Edilanne Katrine Amparo Viana, Hellen Cris Araújo Souza, Maycon Douglas Araújo Souza, Arthur Serejo Neves Ribeiro, Vanessa de Sousa do Vale, Muhammad Torequl Islam, Joabe Lima Araújo, and Jefferson Almeida Rocha. 2023. "Bioactive Components of Myracrodruon urundeuva against SARS-CoV-2: A Computational Study" Drugs and Drug Candidates 2, no. 4: 781-795. https://doi.org/10.3390/ddc2040039
APA StyleAlves, S. K. S., Sousa, C. S., Viana, E. K. A., Souza, H. C. A., Souza, M. D. A., Ribeiro, A. S. N., Vale, V. d. S. d., Islam, M. T., Araújo, J. L., & Rocha, J. A. (2023). Bioactive Components of Myracrodruon urundeuva against SARS-CoV-2: A Computational Study. Drugs and Drug Candidates, 2(4), 781-795. https://doi.org/10.3390/ddc2040039