Small Ruminant Genetics and Breeding
A topical collection in Animals (ISSN 2076-2615). This collection belongs to the section "Small Ruminants".
Viewed by 51010Editor
Interests: animal genetics; aquaculture and fisheries genetics; quantitative genetics; population and conservation genetics; molecular genetics
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
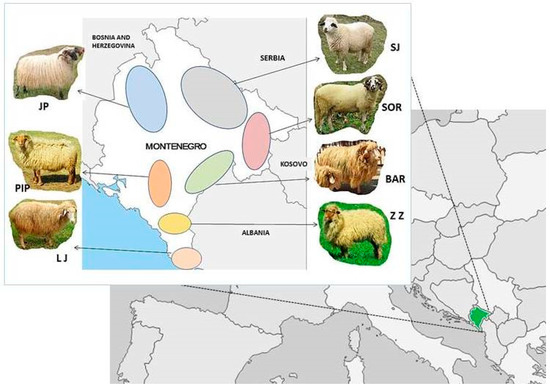
Small ruminants, such as sheep (Ovis aries) and goats (Capra hircus), were among the first animals to be domesticated, with historical evidence linking them to Western Asia approximately 9000–12,000 years ago. Domesticated sheep and goats provided early humans with a supply of fiber, pelt, meat, and milk. Owing to their small stature and versatility, small ruminants have become steadily important in the global rural economy, especially in the arid and semi-arid regions. Furthermore, the demand for meat and milk in developing countries is constantly increasing, and a sustainable increase in small ruminant production would therefore be desirable in order to meet the demands of the human population on livestock populations and their products.
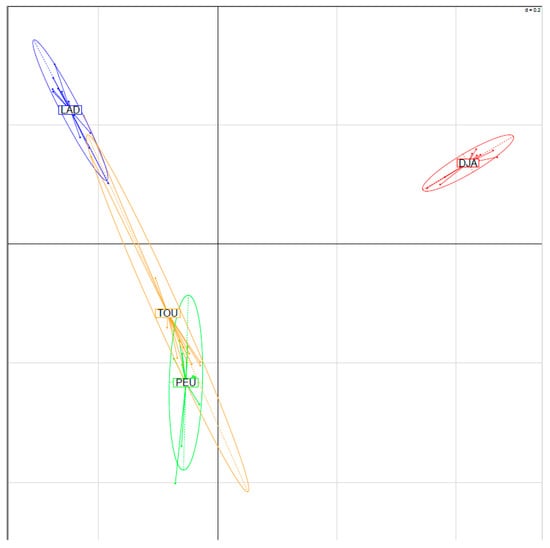
Genetic improvement can substantially promote the efficiency of animal production, by increasing the performance of small ruminant flocks or populations over time with the use of genetically superior animals. A prerequisite is to select the most desirable breed or breed combination, and to define the breeding objectives.
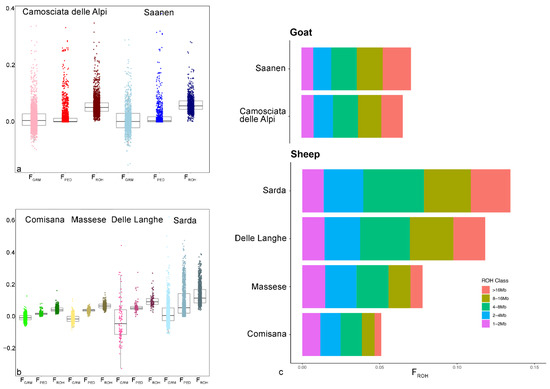
Over the last two decades, advances in DNA technology have dramatically increased the efficiency and the affordability of gaining genome information, leading to the development of fast, cost-effective, and more accurate methods for the implementation of breeding programs.
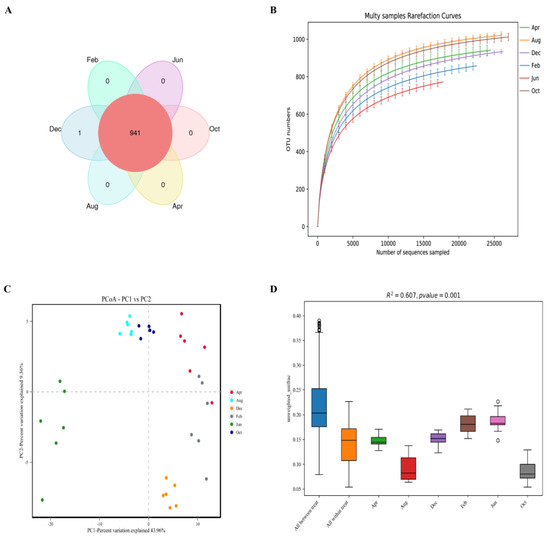
For this Special Issue, original research manuscripts covering all aspects of small ruminant genetics, such as population genetics, breed investigation and characterization, quantitative genetics, QTL and marker assisted selection, genomic selection, gene polymorphisms, and genome-wide association studies, are welcome.
Dr. Dimitrios Loukovitis
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Animals is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2400 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- small ruminant
- genetic improvement
- population genetics
- QTL
- genomic selection
- molecular markers