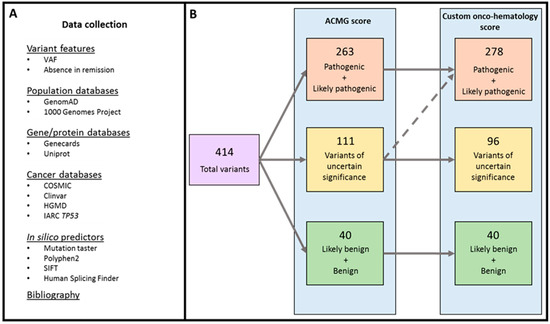
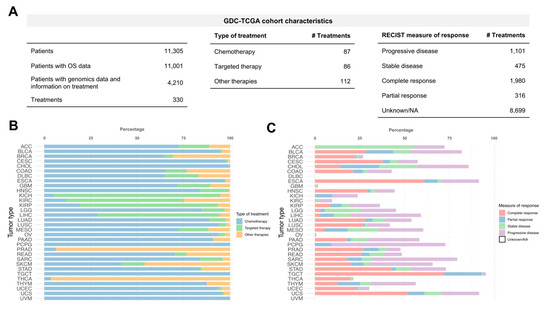
Application of Bioinformatics in Cancers
A topical collection in Cancers (ISSN 2072-6694). This collection belongs to the section "Cancer Informatics and Big Data".
Viewed by 239670Editor
Interests: functional genomic; proteomic and bioinformatics approaches in cancer; sequencing the exomes and transcriptomes of head and neck cancer; drug sensitivities
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
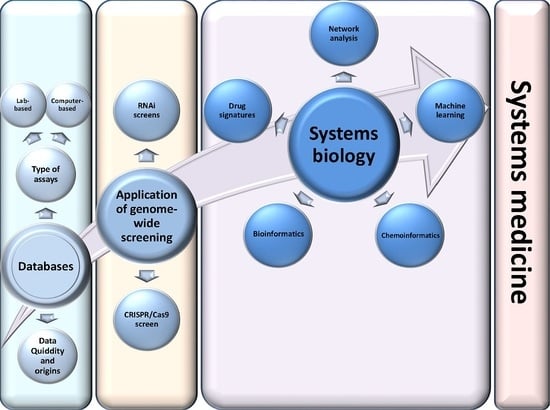
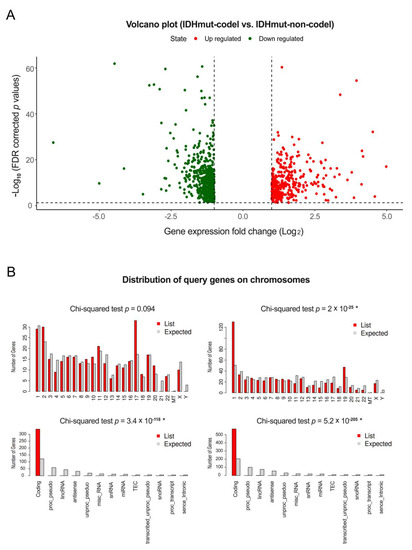
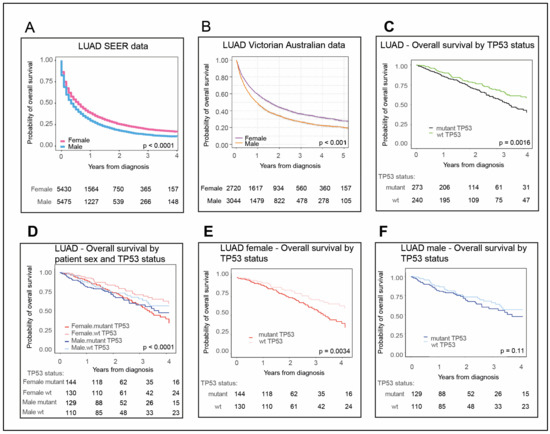
Bioinformatic applications in cancer have rapidly evolved over the past several years. Ever since its initial implementation, next generation sequencing has altered our understanding of cancer biology, and the approaches to analyzing the more and more complex datasets have also become increasingly complex. Routine bioinformatics pipelines now range from those that rapidly detect and predict the functional impact of molecular alterations, to those that quantify the changes in the tumor microenvironment. For example, several tools that analyze tumor–immune interactions have been successfully developed so as to assess the tumor infiltrating lymphocyte content, microsatellite instability, total mutational burden, and neoantigen presentation. The further complexity of the integrated omics-based analysis is also now coupled with the emergence of modern machine learning and network-based approaches for analyzing large datasets in the context of publicly available resources, such as the cancer genome atlas.
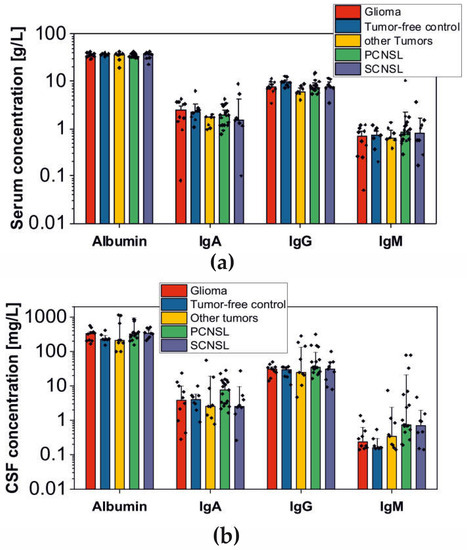
While much of the focus has so far been on annotating molecular alterations. as well as infiltrating cell types or cell states in ideal sequencing conditions, alternative and application-specific approaches are now emerging that improve on a wide variety of established analysis techniques. These include techniques that range from the improved quantification of the copy number and gene expression from formalin-fixed tissues. as well as applications that require a high sensitivity, such as the quantification of tumor mutations from liquid biopsies (circulating cell free DNA). Further novel applications attempt to improve the ability to analyze the distribution and molecular impact of complicated genetic features, such as repetitive or transposable endogenous elements (e.g., LINE-1), as well as exogenous genetic elements (e.g., human papilloma virus).
As we develop a better understanding of the limitations of these new informatics approaches, we can ultimately hope to apply these techniques to the existing datasets, and build well-annotated databases of easily accessible information that can be leveraged in multi-variable analysis pipelines. Similar to the success of SIGdb and cBioPortal, this should help yield new diagnostic and prognostic/predictive biomarkers for standard interventional modalities, as well as emerging areas like immuno-oncology, and areas of unmet clinical need. This Topical Collection will highlight the current state-of-the-art in bioinformatics applications in cancer biology, and infer future prospects for improving informatics applications through artificial intelligence and machine learning approaches.
Dr. J. Chad Brenner
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cancers is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2900 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.