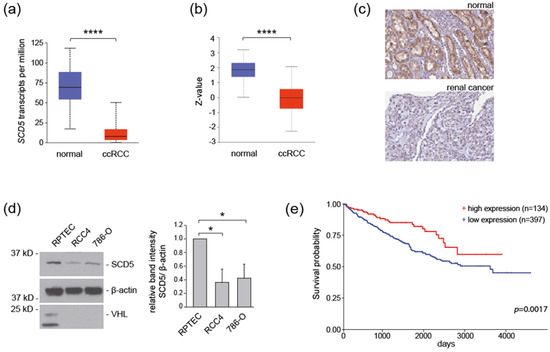
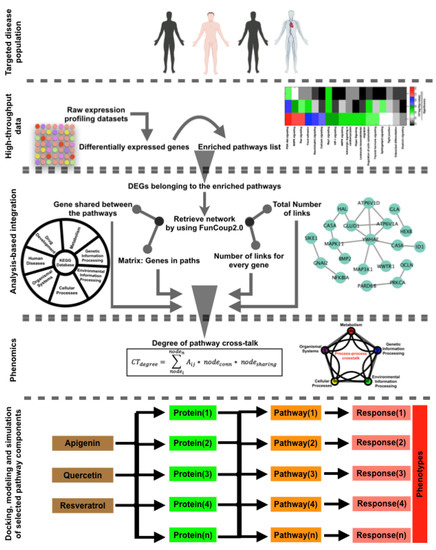
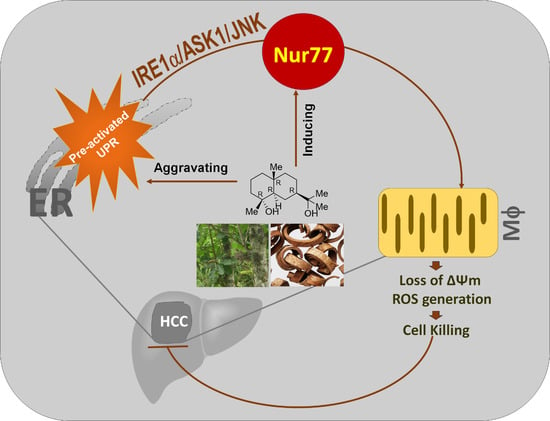
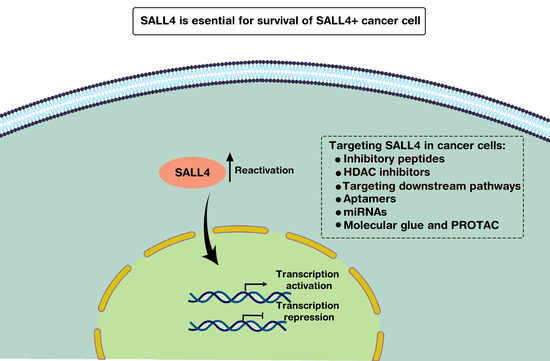
Emerging Cancer Target Genes
A topical collection in Cells (ISSN 2073-4409). This collection belongs to the section "Cell and Gene Therapy".
Viewed by 136096Editor
Interests: acute myeloid leukemia; cell biology; metabolism; molecular biology; genetics; gene editing
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
With the development of a multitude of new high-throughput techniques in recent years, basic research has reached an unprecedented pace with the result that an incredible amount of data can be produced year by year, which far exceeds the capacity of researchers to fully validate it. This includes identifying a large number of candidate target genes that could potentially be used in therapy for the treatment of patients.Novel therapeutic options are urgently sought, particularly in cancer research, which very often focuses on entities in which diagnosis means a highly restricted quality of life and greatly shortened survival. This Topical Collection of Cells invites contributions that have experimentally identified such new targets, such as novel oncogenes or tumor suppressor genes, cancer-inducing or -repressing long noncoding RNAs or cellular signaling and metabolic pathways influencing tumorigenesis. Additionally, we are interested in new therapeutic options, such as new drugs or drugs with improved bioavailability, drug combinations with proven synergistic activity or new innovative strategies to fight tumors. The identification of each of these targets offers the opportunity to develop and refine therapies, or to combine therapies to achieve the greatest possible success for the patient.
Dr. Frank Schnütgen
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- Cancer
- Cancer therapy
- Genetic screening
- Therapeutic targets
- Metabolism
- Transcriptomics