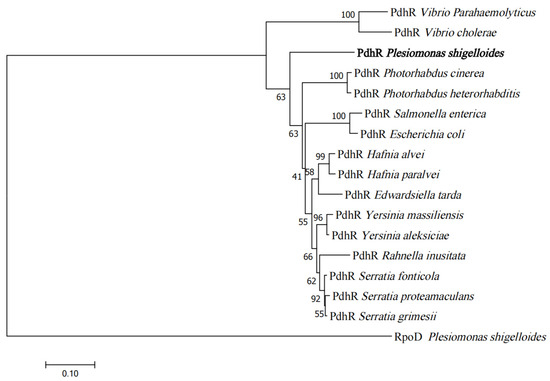
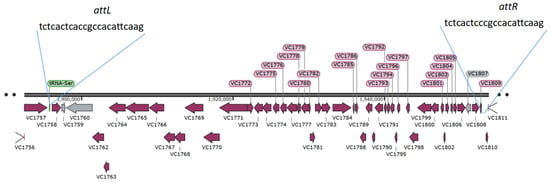
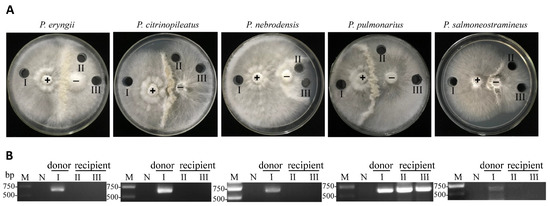
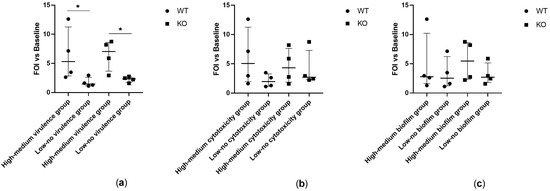
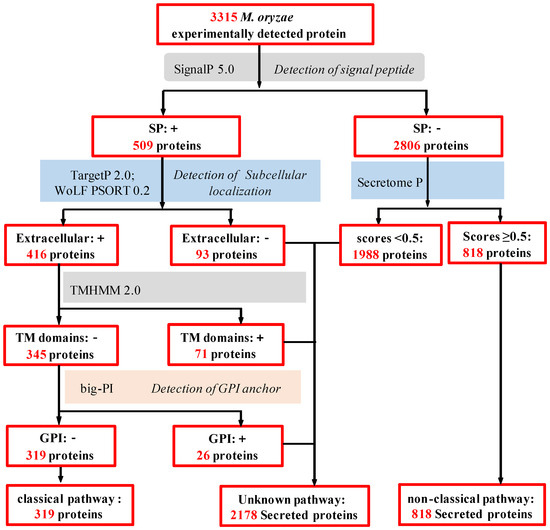
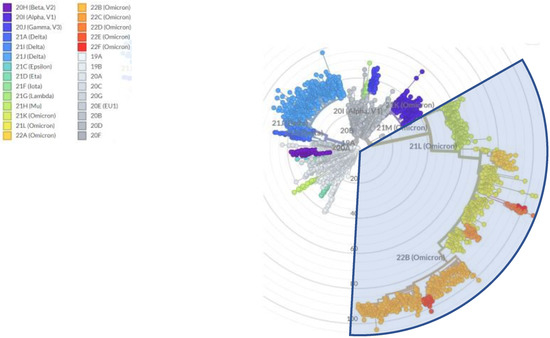
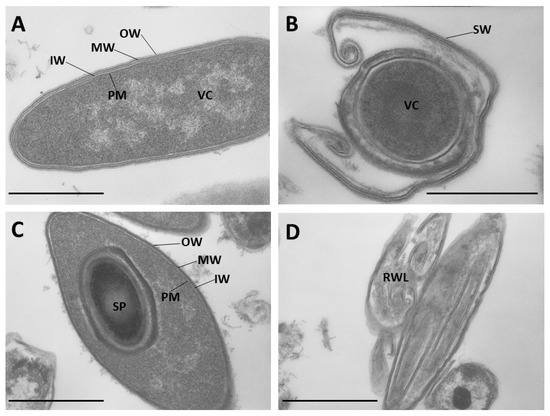
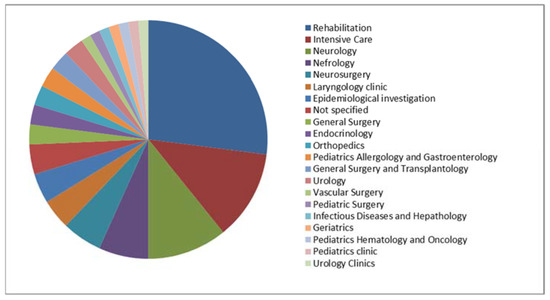
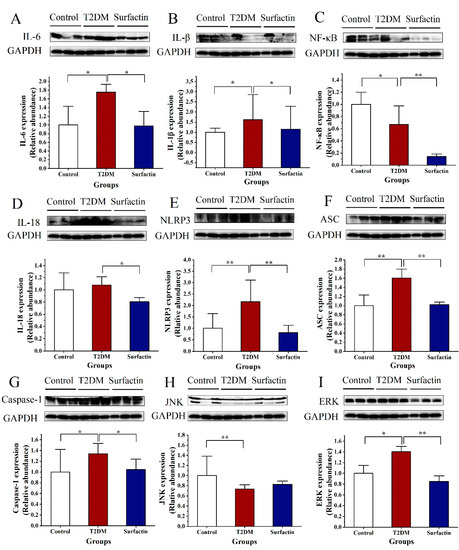
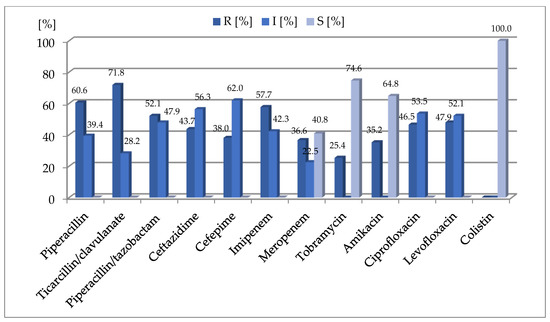
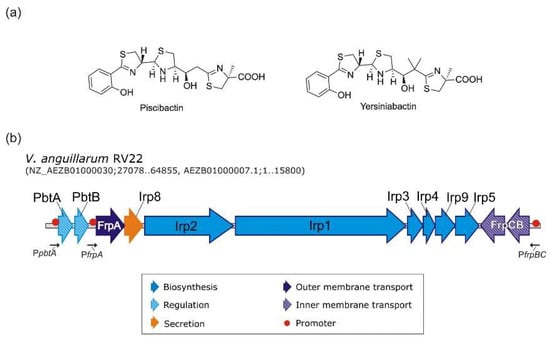
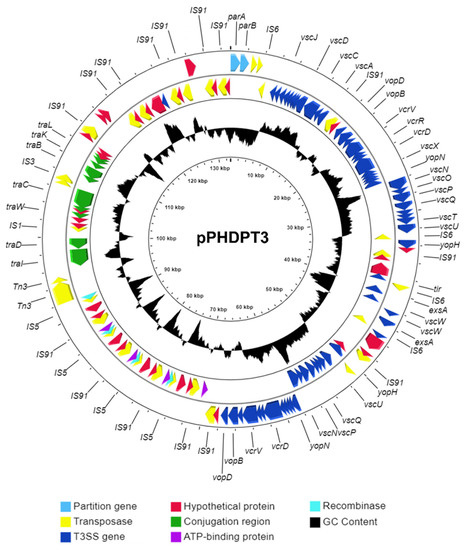
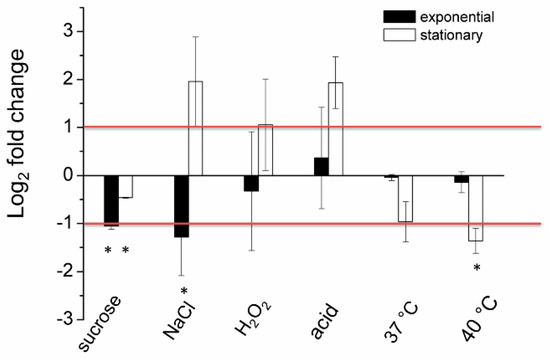
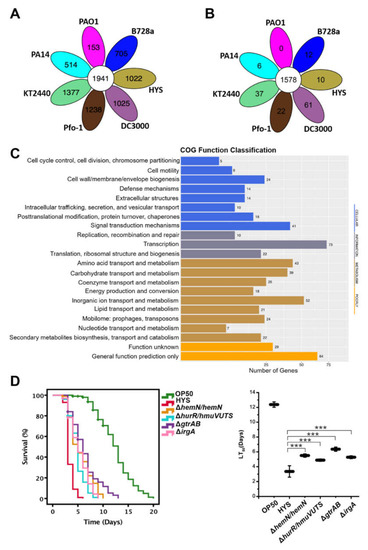
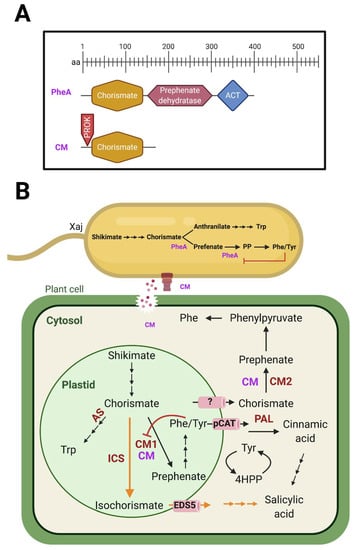
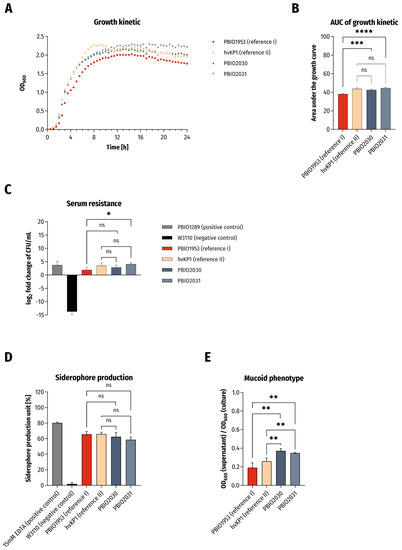
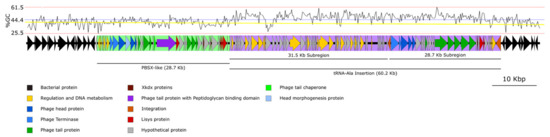
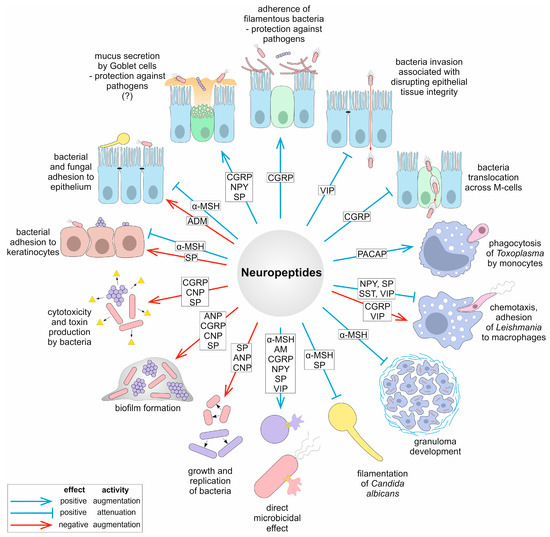
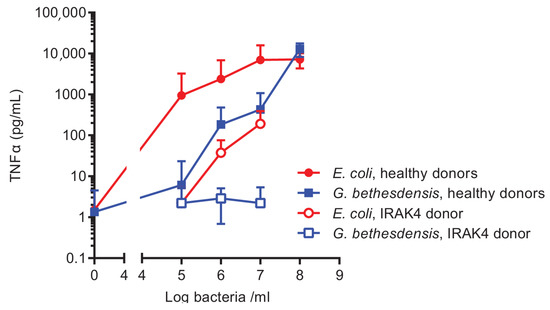
Microbial Virulence Factors
A topical collection in International Journal of Molecular Sciences (ISSN 1422-0067). This collection belongs to the section "Molecular Microbiology".
Viewed by 67956Editor
Interests: molecular microbiology; biology and biochemistry of Gram-negative bacteria; bacterial small non-coding regulatory RNAs; mechanisms of resistance to antimicrobials; development of new antimicrobials; vaccine research
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
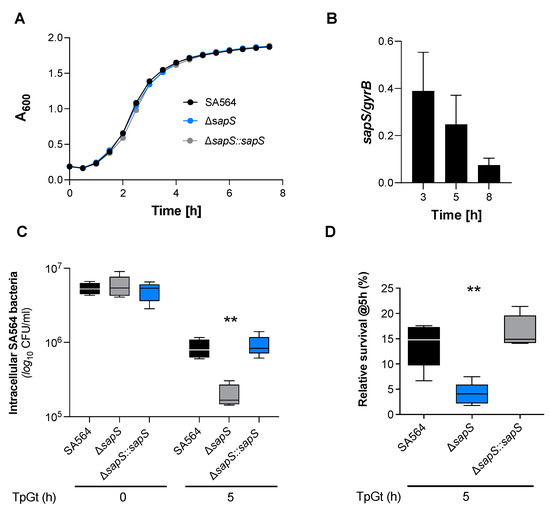
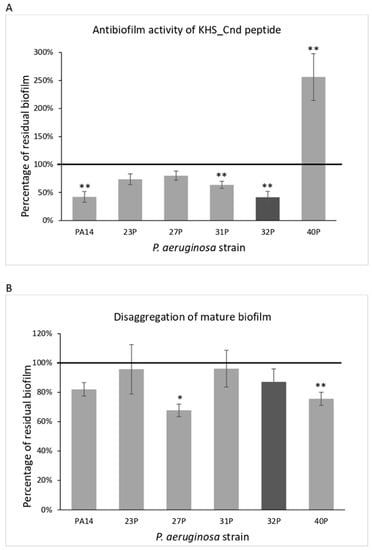
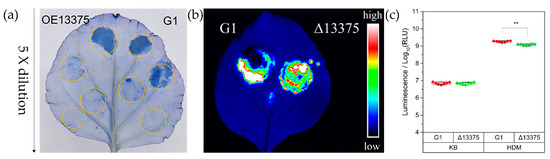
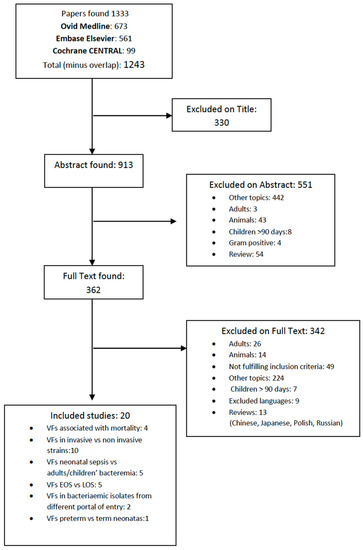
Microbial virulence factors can be defined as a wide range of molecules produced by pathogenic microbes that enhance their ability to evade their host defenses and cause disease. This broad definition encompasses secreted products such as toxins, enzymes, and exopolysaccharides, but also cell surface structures such as capsules, lipopolysaccharides, glyco- and lipoproteins, and intracellular changes in metabolic regulatory networks governed by protein sensors/regulators and noncoding regulatory RNAs. The knowledge, at a molecular level, of the biology of microbial pathogens and their virulence factors is central in the development of novel therapeutic molecules and strategies to combat microbial infections. This is of particular importance at present with the worldwide emergence of microbes resistant to available antimicrobials. Advances in recent years in molecular biology, genomic technologies, and bioinformatics have contributed to the molecular identification and functional analyses of microbial virulence factors. This Topical Collection of IJMS will be focused on virulence factors and their regulatory networks from microbes such as bacteria, viruses, fungi, and parasites, as well as on the description of innovative experimental techniques to characterize microbial virulence factors. Research papers, up-to-date review articles, and commentaries are all welcome.
Prof. Jorge H. Leitão
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. International Journal of Molecular Sciences is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. There is an Article Processing Charge (APC) for publication in this open access journal. For details about the APC please see here. Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- Pathogenic bacteria
- Toxins
- Bacterial capsules
- Fungal virulence factors
- Virus virulence factors
- Host defense evasion
- Intracellular survival
- Virulence determinants
- Human microbial parasites