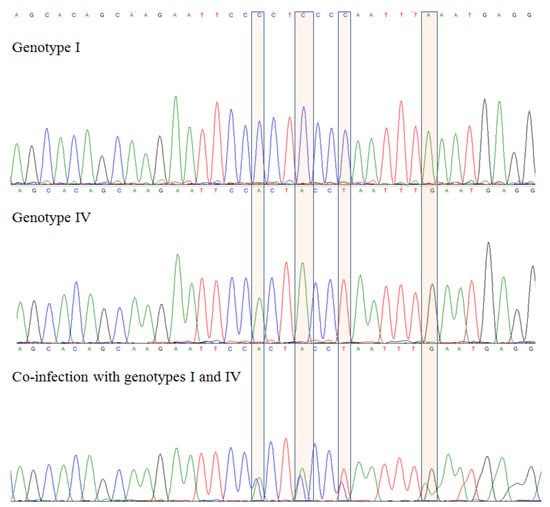
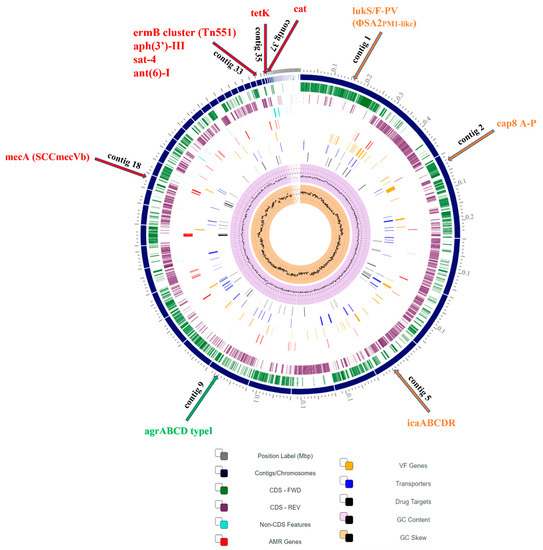
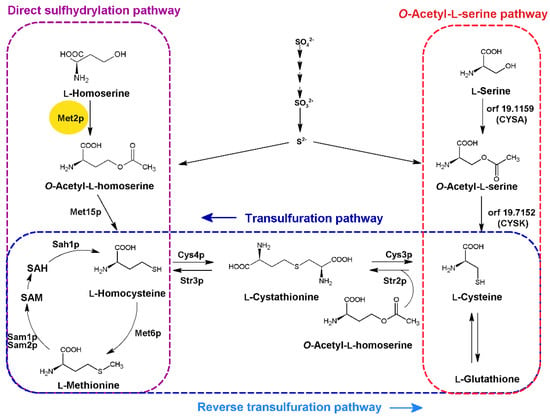
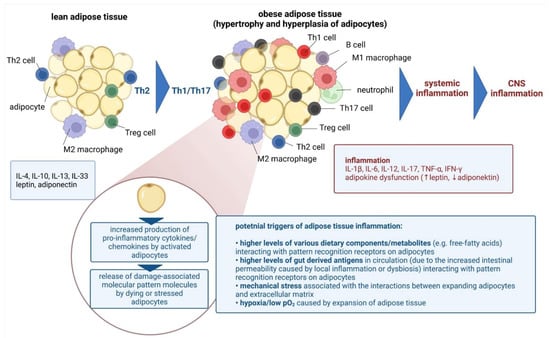
State-of-the-Art Molecular Microbiology in Poland (Closed)
A topical collection in International Journal of Molecular Sciences (ISSN 1422-0067). This collection belongs to the section "Molecular Microbiology".
Viewed by 89365Editors
Interests: biology of bacteriophages; biodiversity of bacteriophages; regulation of bacteriophage development; regulation of phage gene expression; control of phage DNA replication; phage therapy; phages bearing genes of toxins; bacteriophage genomics
Special Issues, Collections and Topics in MDPI journals
Interests: protein folding; heat shock response; peptidyl prolyl cis/trans isomerases; disulfide bond formation; RpoE sigma factor; two-component systems; envelope stress; transcription factors; lipopolysaccharide
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
This Special Issue aims to provide a comprehensive overview of recent advances in molecular microbiology in Poland by inviting contributions from Polish research institutes/laboratories that consolidate our understanding of this area. Topics include, but are not limited to, the following:
- Antibiotic resistance mechanisms;
- Biosynthesis of macromolecules;
- Cell division and cell wall structure;
- Gene expression and its regulation;
- Gene transfer mechanisms;
- Host–pathogen interactions including host responses;
- Induction of cell death by microorganisms;
- Membrane biogenesis and function;
- Pathogenicity mechanisms;
- Posttranslational modifications;
- Protein delivery (secretion and trafficking);
- Signaling pathways and networks;
- Systems biology;
- Vaccines;
- Virulence factors.
Dr. Alicja Wegrzyn
Prof. Dr. Satish Raina
Guest Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. International Journal of Molecular Sciences is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. There is an Article Processing Charge (APC) for publication in this open access journal. For details about the APC please see here. Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- molecular microbiology
- gene expression regulation
- biochemical processes
- pathogenicity mechanisms
- host-microbe interactions
- bacteriophages
Planned Papers
The below list represents only planned manuscripts. Some of these manuscripts have not been received by the Editorial Office yet. Papers submitted to MDPI journals are subject to peer-review.
Title 1: Complex genome destabilization followed by compensatory evolution in mitotically dividing yeast cells
Title 2: The yeast histone chaperone Asf1 down-regulates protein kinase CK2
Title 3:Genetic diversity of satelite RNAs of tomato black ring virus and their impact on virus replication
Title 4: An approach for catalytically active serine proteases isolation with biotinylated aminophosphonates
Title 5: Molecular mechanisms of drug resistance in Staphylococcus aureus
Title 6: Mechanisms of antibiotic resistance and molecular epidemiology of Neisseria gonorrhoeae