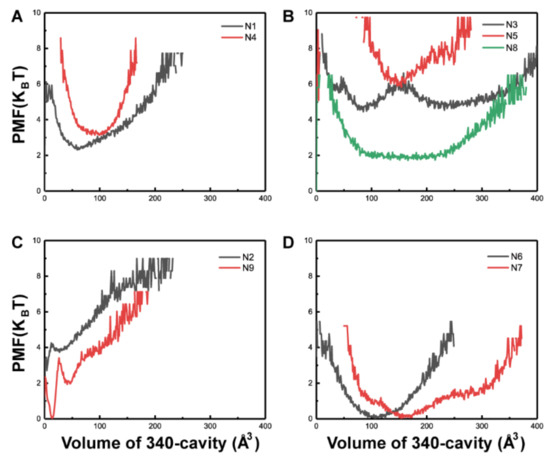
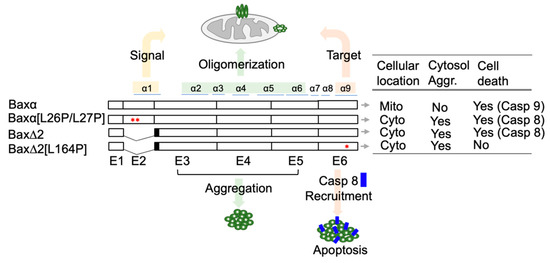
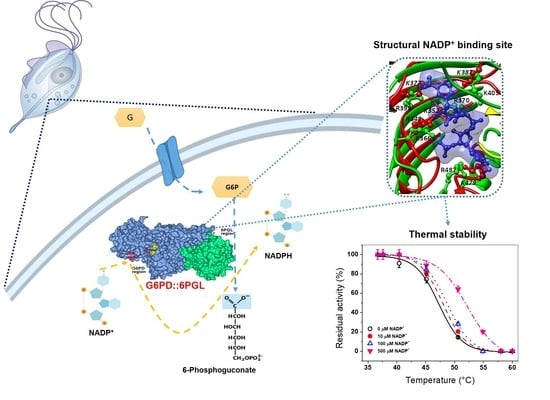
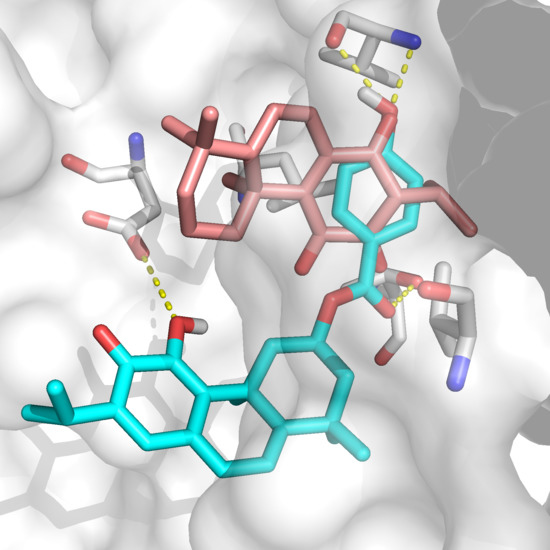
Computational Studies of Biomolecules (Closed)
A topical collection in International Journal of Molecular Sciences (ISSN 1422-0067). This collection belongs to the section "Molecular Biophysics".
Viewed by 152883Editors
Interests: enzyme reaction mechanisms and dynamics; non-heme iron histone demethylases; multiscale modeling of epigenetic mechanisms
Special Issues, Collections and Topics in MDPI journals
Interests: computational chemical biology; enzyme mechanisms; catalytic activity and inhibition; computer-aided drug design; conformational dynamics of proteins and nucleic acids; biomolecular spectroscopy; bioinorganic enzymology
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
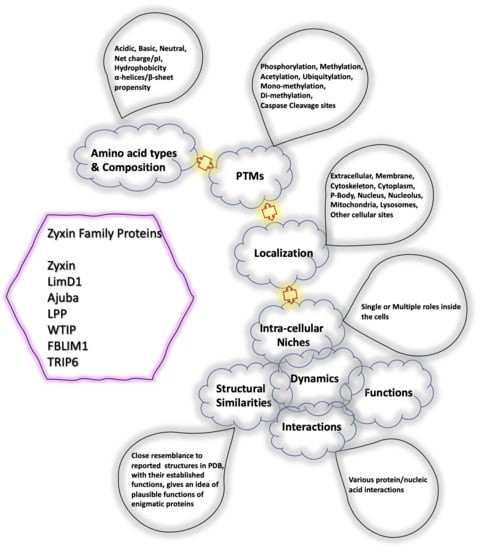
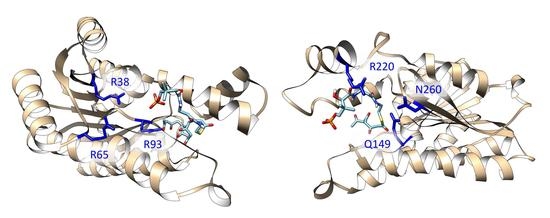
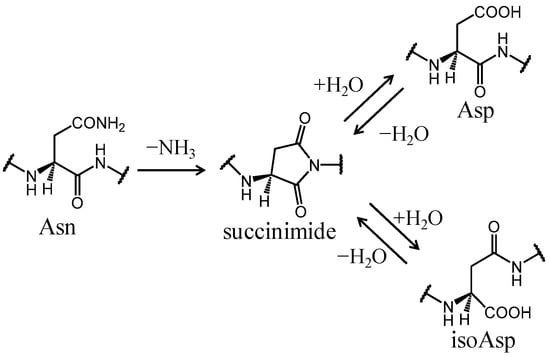
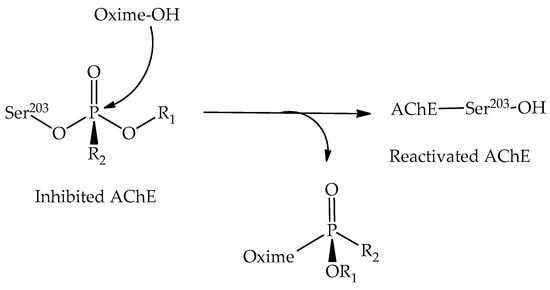
Computational chemistry methods are nowadays widely applied for studying biomolecular structure, mechanisms, dynamics, and function. Molecular dynamic (MD) simulation methods, quantum mechanic (QM) methods, combined quantum mechanics/molecular mechanics (QM/MM), molecular docking, and other computational techniques have proven to be very useful for fundamental understanding of structure–function relationships in biomolecules, but also very useful for applications in drug design, chemical biology, and biotechnology. Importantly, the increased computational power and the development of high-performance computing have made it possible to achieve growth in synergistic computational–experimental studies in the most topical areas of biomolecular sciences in a timely manner.
The current Topical Collection aims to attract high-quality contributions of modeling biomolecular structures, dynamics, functions, and interactions with the potential of interpretation of experimental data and applications in drug design and protein design.
Topics of interest:
- Development and validation of new computational modeling methods;
- Computational studies of proteins’ structure–function relationships;
- Computational investigations of nucleic acids’ structure–function relationships;
- Modeling of protein and nucleic acid dynamics;
- Protein docking;
- Protein–ligand interactions;
- Nucleic acid–ligand interactions;
- Protein design;
- Computational enzymology–enzymatic reaction mechanisms;
- Protein homology modeling.
Prof. Dr. Christo Z. Christov
Dr. Tatyana Karabencheva-Christova
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. International Journal of Molecular Sciences is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. There is an Article Processing Charge (APC) for publication in this open access journal. For details about the APC please see here. Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.