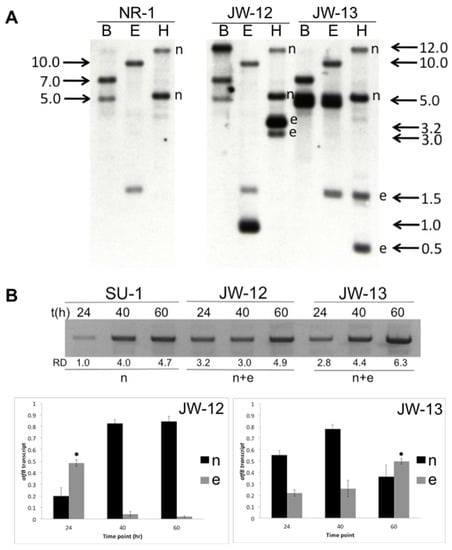
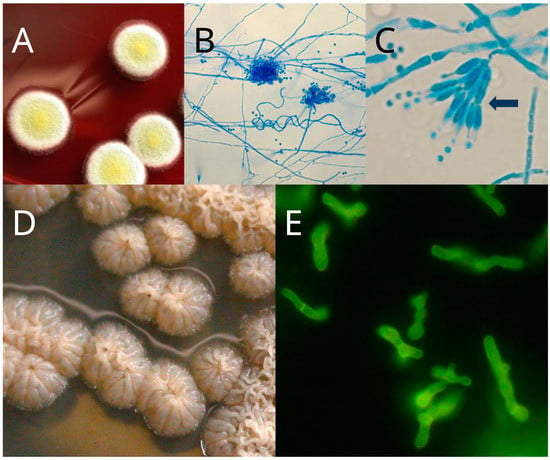
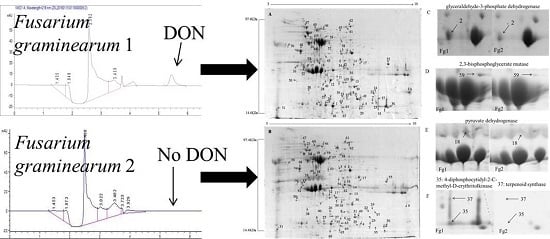
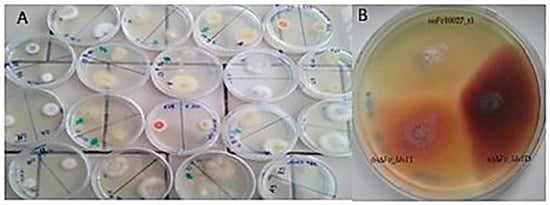
Evolutionary/Phylogenetic Studies of Mycotoxin Biosynthetic Pathways
A topical collection in Toxins (ISSN 2072-6651). This collection belongs to the section "Mycotoxins".
Viewed by 133202Editors
Interests: regulation and biosynthesis of mycotoxins; fungal endophyte-grass relationships; bacterial endophytes, and the coevolution of secondary products; primarily mycotoxins, with grasses and other plants, as an adaptive strategy for mutualistic associations
Special Issues, Collections and Topics in MDPI journals
Special Issues, Collections and Topics in MDPI journals
Interests: aflatoxins; mycotoxins; Aspergillus flavus; genomics; functional genomics; Next generation sequencing (NGS); plant-fungus interaction; genomics of Penicillium spp and their secondary metabolites
Topical Collection Information
Dear Colleagues,
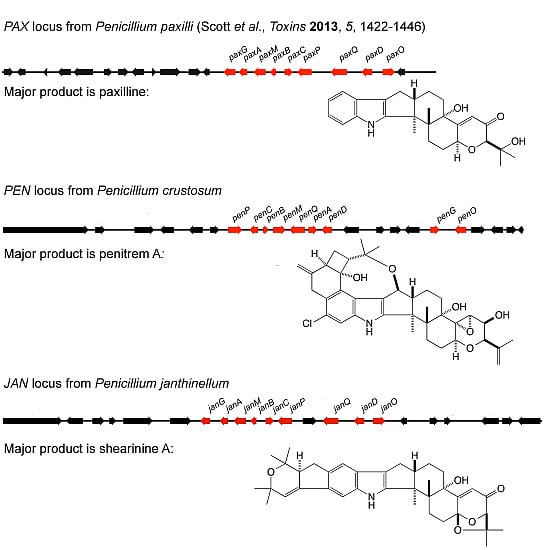
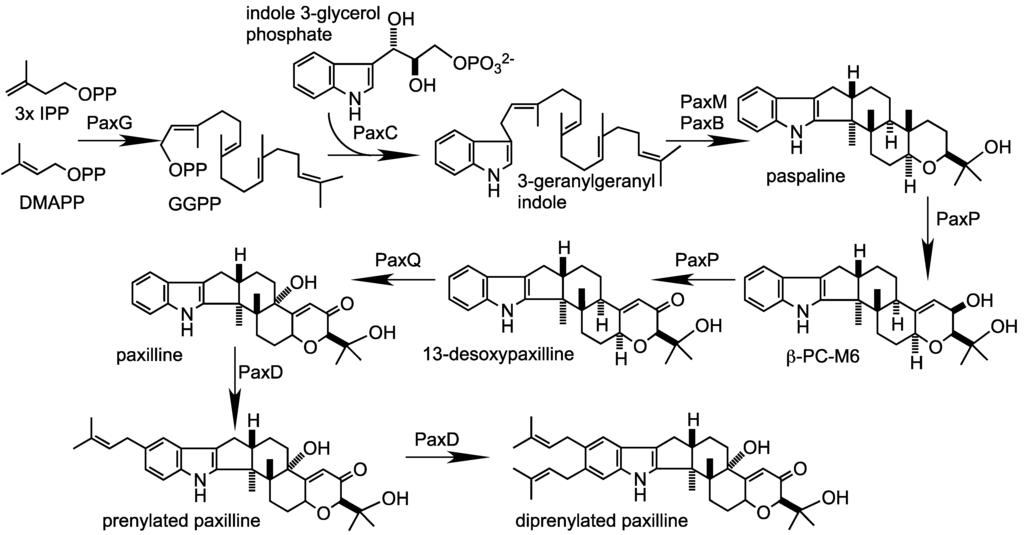
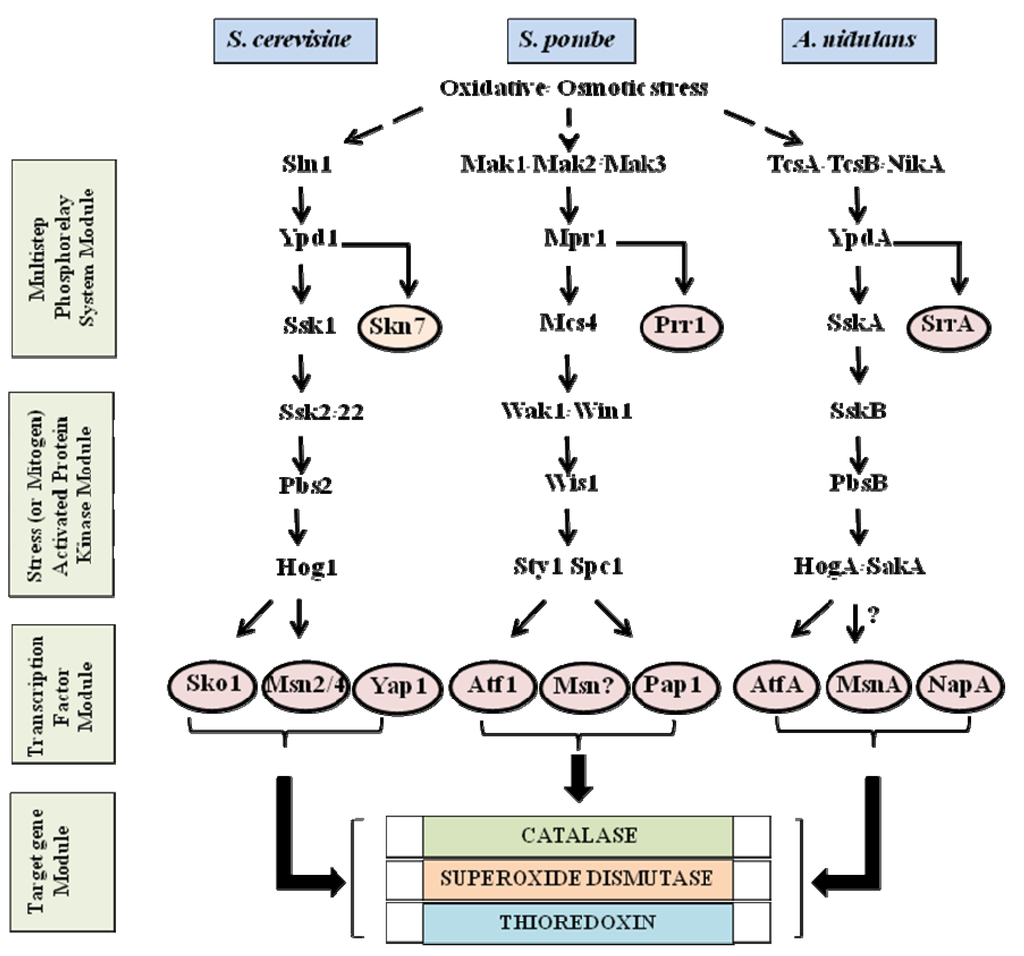
In the fungal kingdom, many species are capable of producing a variety of secondary metabolites. Some are beneficial and have pharmaceutical properties, while quite a number of secondary metabolites are toxic and carcinogenic to animal and human beings. These harmful metabolites are known as mycotoxins. Well-studied mycotoxins include aflatoxins, ochratoxins, sterigmatocystins, cyclopiazonic acid (CPA), kojic acid, patulin, citrinin, fumonisins, trichothecenes, deoxynivalenol (DON) toxins, and zearalenone toxin etc. are produced from Penicillium, Aspergillus, Fusarium and other fungal species.
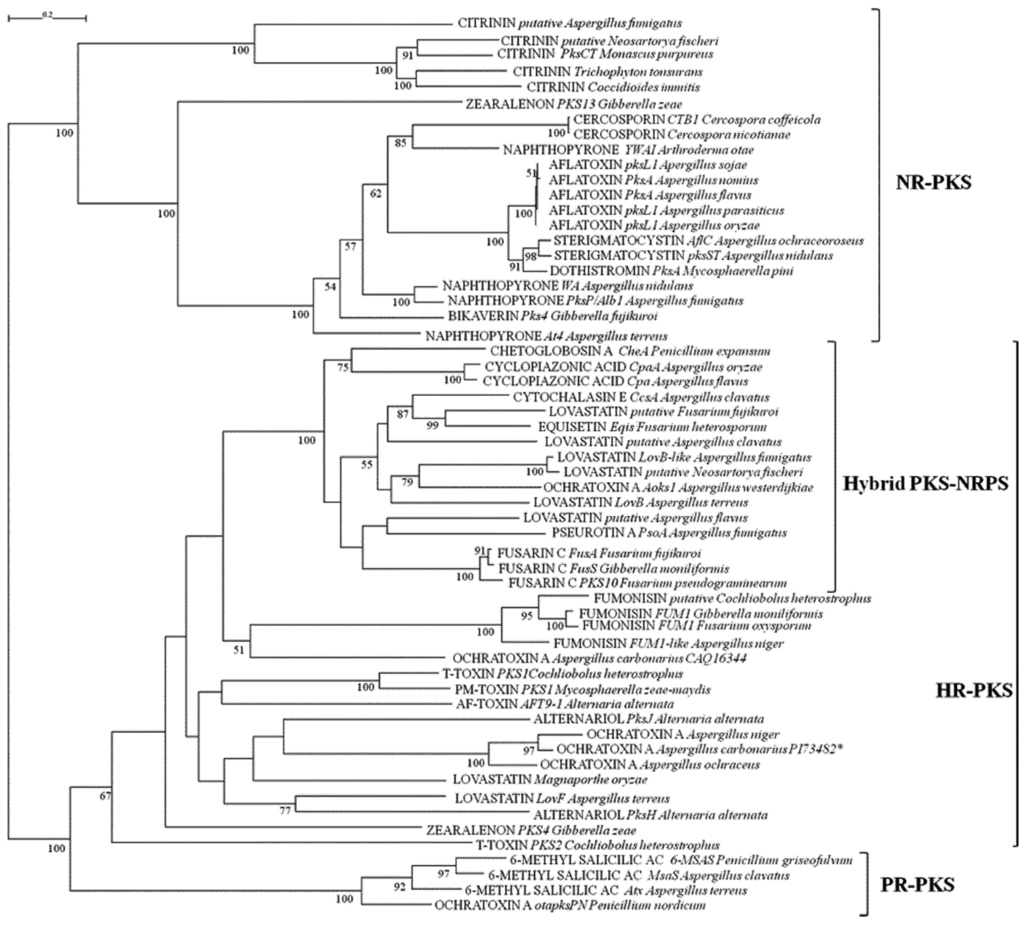
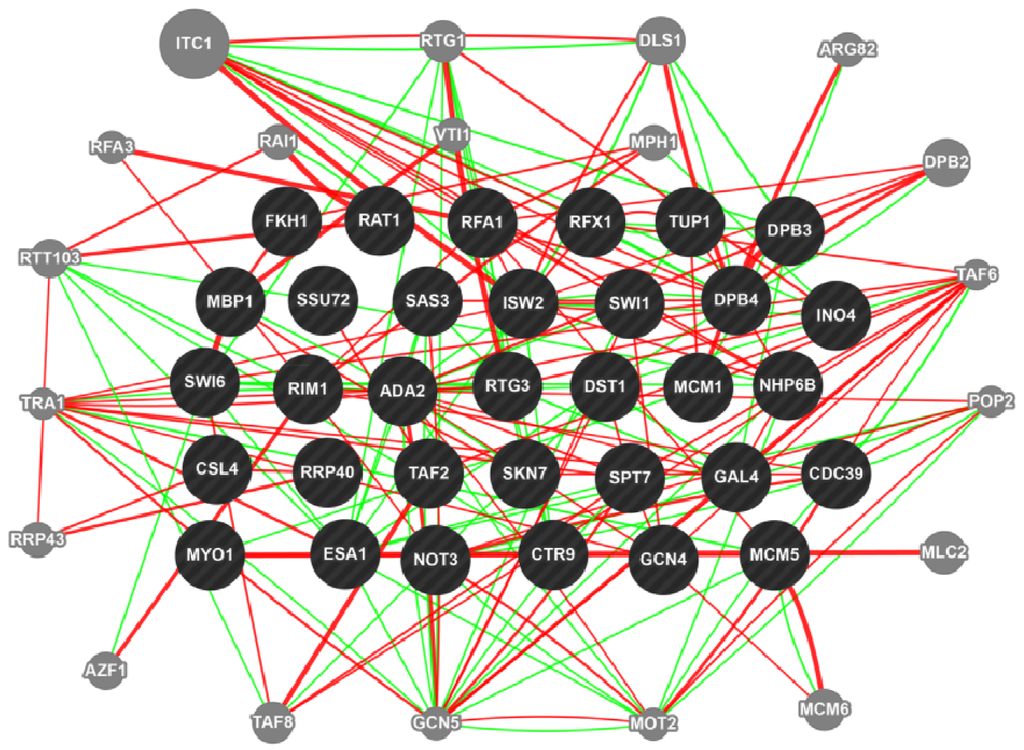
Biosynthetic pathways, pathway gene clusters, and their genetic regulation have been widely reported. The genomes of most fungal and related species have been sequenced. In the post-genomic era, genes and gene clusters within those genomes have been investigated. Comparative genomics analysis of related toxigenic fungal species has revealed a vast reservoir of information concerning mycotoxin biosynthetic pathways, and their pathway gene clusters, as well as their organization in the genome. This sequencing data will enrich our knowledge of the evolutionary statuses and phylogenetic relationships of related fungal species. This Special Issue aims to better our understanding of the evolutionary processes and phylogenetic relationships of biosynthetic pathways, and their pathway gene and gene clusters in related genomes. We expect that this knowledge can be translated into biotechnological strategies for preventing mycotoxin contamination in food and feed. Topics of this Special Issue on “Evolutionary/Phylogenetic Studies of Mycotoxin Biosynthetic Pathways” include:
- Comparative and evolutionary genomics
- Phylogenetic relationships of mycotoxigenic fungi
- Evolution of secondary metabolism gene clusters
- Horizontal gene transfer in secondary metabolism gene cluster evolution
- Interactions between mycotoxigenic fungi on mycotoxin biosynthesis
- Bioinformatic methods in the analysis of fungal evolution
Dr. Jiujiang Yu
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a double-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Toxins is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- mycotoxigenic fungi
- evolution
- horizontal gene transfer
- phylogenic relations
- secondary metabolism
- biosynthetic pathways
- pathway gene clusters
- food safety
- feed safety
Related Special Issue
- Evolutionary/Phylogenetic Studies of Mycotoxin Biosynthetic Pathways 2013 in Toxins (7 articles - displayed below)