Structure of Supramolecular Assemblies Formed by α,δ-Tetramethylcucurbit[6]uril and 4-Nitrophenol
Abstract
:Introduction

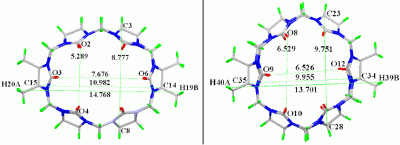
Results and Discussion



| C-H…π | C-H(Å) | H…Cg(Å) | C…Cg(Å) | C-H…Cg(Å) | aromatic ring | symmetry codes |
|---|---|---|---|---|---|---|
| C46-H46…Cg(1) | 0.93 | 2.862 | 3.647(3) | 120.27 | C65-C70 | |
| C67-H67…Cg(2) | 0.93 | 2.802 | 3.629(3) | 148.80 | C71-76 | x, y, 1+z |
| C75-H75…Cg(3) | 0.93 | 2.620 | 3.316(4) | 132.14 | C59-C64 | |
| C63-H63…Cg(4) | 0.93 | 2.733 | 3.533(3) | 144.68 | C41-C46 | x, y, -1-z |
| C57-H87…Cg(5) | 0.93 | 3.693 | 4.332(4) | 128.03 | C83-C88 | |
| C87-H87…Cg(6) | 0.93 | 2.985 | 3.601(3) | 125.10 | C77-C82 | |
| C79-H79…Cg(7) | 0.93 | 2.942 | 3.499(3) | 119.81 | C47-C52 | |
| C49-H49…Cg(8) | 0.93 | 2.645 | 3.696(3) | 152.80 | C53-C58 | 1-x, 1-y, 1-z |


Experimental
General
Preparation of (TMeQ[6]-4nitrophenol)·13H2O (1)
X-ray Crystallography
| Empirical formula | C88H110N32O49 | Dcalcd, g cm-3 | 1.491 |
| Formula weight | 2400.08 | T, K | 223 |
| Crystal system | Monoclinic | μ, mm-1 | 0.123 |
| Space group | P-1 | unique reflns | 18398 |
| a, Å | 17.758(6) | obsdreflns | 11799 |
| b, Å | 17.908(6) | params | 1533 |
| c, Å | 18.941(7) | Rint | 0.0430 |
| α,deg | 88.290(5) | R[I> 2σ(I)]* | 0.0828 |
| β,deg | 83.072(5) | wR[I> 2σ(I)]# | 0.2562 |
| γ,deg | 63.602(5) | R(all data) | 0.1228 |
| V, Å3 | 5346 | WR(all data) | 0.2786 |
| Z | 2 | GOF on F2 | 1.052 |
Acknowledgements
Supporting Information Available
References
- Lagona, J.; Mukhopadhyay, P.; Chakrabarti, S.; Isaacs, L. The Cucurbit[n]uril Family. Angew. Chem. Int. Ed. 2005, 44, 4844. [Google Scholar] [CrossRef]
- Freeman, W. A.; Mock, W. L.; Shih, N. Y. Cucurbituril. J. Am. Chem. Soc. 1981, 103, 7367–7368. [Google Scholar] [CrossRef]
- Day, A. I.; Arnold, A. P.; Blanch, R. J. Method for Synthesis of Cucurbiturils. WO Patent 0068232, 2000. [Google Scholar]
- Kim, J.; Jung, I. S.; Kim, S. Y.; Lee, E.; Kang, J. K.; Sakamoto, S.; Yamaguchi, K.; Kim, K. New Cucurbit Homologues: Syntheses, Isolation, Characterization, and X-ray Crystal structures of Cucurbit[n]uril (n= 5, 7,and 8). J. Am. Chem. Soc. 2000, 122, 540–541. [Google Scholar]
- Day, A. I.; Blanck, R. J.; Amold, A. P.; Lorenzo, S.; Lewis, G. R.; Dance, I. A cucurbituril-based gyroscane: a new supramolecular form. Angew. Chem. Int. Ed. 2002, 41, 275–277. [Google Scholar]
- Zhao, J.; Kim, H. J.; Oh, J.; Kim, S. Y.; Lee, J.; Sakamoto, W. S.; Yamaguchi, K.; Kim, K. Cucurbit[n]uril Derivatives Soluble in Water and Organic Solvents. Angew. Chem. Int. Ed. 2001, 40, 4233–4235. [Google Scholar] [CrossRef]
- Sasmal, S.; Sinha, M. K.; Keinan, E. Facile Purification of Rare Cucurbiturils by Affinity Chromatography. Org. Lett. 2004, 6, 1225–1228. [Google Scholar] [CrossRef]
- Jon, S. Y.; Selvapalam, N.; Oh, D. H.; Kang, J. K.; Kim, S. Y.; Jeon, Y. J.; Lee, J. W.; Kim, K. Facile Synthesis of Cucurbit[n]uril Derivatives via Direct Functionalization: Expanding Utilization of Cucurbit[n]uril. J. Am. Chem. Soc. 2003, 125, 10186–10187. [Google Scholar]
- Isobe, H.; Sato, S.; Nakamura, E. Synthesis of disubstituted cucurbit[n]uril and its rotaxane derivative. Org. Lett. 2002, 4, 1287–1289. [Google Scholar] [CrossRef]
- Day, A. I.; Arnold, A. P.; Blanch, R. J. A method for synthesizing partially substituted cucurbit[n]uril. Molecules 2003, 8, 74–84. [Google Scholar] [CrossRef]
- Zhao, Y.-J.; Xue, S.-F.; Zhu, Q.-J.; Tao, Z.; Zhang, J.-X.; Wei, Z.-B.; Long, L.-S.; Hu, M.-L.; Xiao, H.-P.; Day, A. I. Synthesis of a symmetrical tetrasubstituted cucurbit[6]uril and its host-guest compound with 2, 2’-bipyridine. Chin. Sci. Bull. 2004, 49, 1111–1116. [Google Scholar] [CrossRef]
- Lagona, J.; Fettinger, J. C.; Isaacs, L. Cucurbit[n]uril Analogues. Org. Lett. 2003, 5, 3745–3747. [Google Scholar] [CrossRef]
- Fu, H.-Y.; Xue, S.- F.; Zhu, Q.-J.; Tao, Z.; Zhang, J.-X.; Day, A. I. Investigation of host – guest compounds of cucubit[n=5-8]uril with some ortho pyridiniumammonium Ions. J. Inc. Phenom. Macro. Chem. 2005, 52, 101–107. [Google Scholar]
- Kim, K.; Kim, D.; Lee, J. W.; Ko, Y. H.; Kim, K. Growth of poly(pseudorotaxane) on gold using host – stabilized charge – transfer interaction. Chem. Commun. 2004, 848–849. [Google Scholar]
- Pattabiraman, M.; Natarajan, A.; Kaliappan, R. Template directed photodimerization of trans-1,2-bis(n-pyridyl) ethylenes and stilbazoles in water. Chem. Commun. 2005, 4542–4544. [Google Scholar] [CrossRef]
- Buschmann, H. J.; Mutihac, L.; Mutihac, R. C.; Schollmeyer, E. Complexation behavior of cucurbit[6]uril with short polypeptides. Thermochim. Acta 2005, 430, 79–82. [Google Scholar] [CrossRef]
- Lee, H. K.; Park, K. M.; Jeon, Y. J.; Kim, D.; Oh, D. H.; Kim, H. S.; Park, C. K.; Kim, K. Vesicle Formed by Amphiphilc Cucurbit[6]uril: Versatile, Noncovalent Modification of the Vesicle Surface, and Multivalent Bindingof Sugar – Decorated Vesicles to leetin. J. Am. Chem. Soc. 2005, 127, 5006–5007. [Google Scholar]
- Sindelar, V.; Cejas, M. A.; Raymo, F. M.; Kaifer, A. E. Tight inclusion complexation of 2,7-dimethyldiazapyrenium in cucurbit[7]uril. New J. Chem. 2005, 29, 280–282. [Google Scholar] [CrossRef]
- Freeman, W. A. Structures of the p-xylylenediammonium chloride and calcium hydrogensulfate adducts of the cavitand ‘cucurbituril’, C36H36N24O12. Acta Cryst. 1984, B40, 382–387. [Google Scholar] [CrossRef]
- Heo, J.; Kim, S. Y.; Whang, D.; Kim, K. Shape-Induced, Hexagonal, Open Frameworks: Rubidum Ion Complexed Cucurbituril. Angew. Chem. Int. Ed. 1999, 38, 641–643. [Google Scholar] [CrossRef]
- Sokolov, M. N.; Dybtsev, D. N.; Fedin, V. P. Supramolecular compounds of cucurbituril with molybdenum and tungsten chalcogenide cluster aqua complex. Russ. Chem. Bull. 2003, 52, 1041–1060. [Google Scholar] [CrossRef]
- Gerasko, O. A.; Sokolov, M. N.; Fedin, V. P. Mono and polynuclear aqua complexes and cucurbit[6]uril: versatile building blocks for supramolecular chemistry. Pure Appl. Chem. 2004, 76, 1633–1646. [Google Scholar]
- Mu, L.; Xue, S.-F.; Du, Y.; Zhao, Y.-J.; Zhu, Q.-J.; Tao, Z. Interaction Between Three Cucurbiturils and Hydrochloride Salts of 4,4’- Dipyridyl and Its Derivates. Chem. J. Chin. Univ. 2006, 27, 654–659. [Google Scholar]
- Tao, Z.; Zhang, G.-Y.; Luo, X.-Q.; Xue, S.-F.; Zhu, Q.-J.; Jackson, W. G.; Wei, Z.-B.; Long, L.-S. C–H···π interactions in the [Co(N-(2-aminomethylpyridyl)ethylenediamine)(2-amino- methylpyridine)Cl]2+ system: syntheses, 2D NMR, X-ray structures and energy minimizations*1. Inorg. Chim. Acta. 2004, 357, 953–964. [Google Scholar]
- Lin, R.-G.; Tao, Z.; Xue, S.-F.; Zhu, Q.-J.; Jackson, W. G.; Wei, Z.-B.; Long, L.–S. C–H···π interaction in the [Co(N-(2-pyridylmethyl)-1,3-diaminopropane)(2-aminomethylpyridine)Cl]2+ system : syntheses, 2D NMR spectroscopy, X-ray structures and energy minimisation. Polyhedron 2003, 22, 3467–3474. [Google Scholar] [CrossRef]
- Tao, Z.; Zhu, Q.-J.; Jackson, W. G.; Zhou, Z.-Y.; Zhou, X.-G. On the preference for the sym–fac isomer in the [Co(N,N′-bis(2-aminomethyl)amine)(2,2′-bipyridine)Cl]2+ system. Polyhedron 2003, 22, 263–270. [Google Scholar] [CrossRef]
- Sheldrick, G. M. SHELXL-97 Program for the Solution and Refinement of Crystal structures; University of Göttingen: Göttingen, Germany, 1997. [Google Scholar]
- Sample Availability: Contact the authors.
© 2008 by the authors. Licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zheng, L.-M.; Zhang, Y.-Q.; Zeng, J.-P.; Qiu, Y.; Yu, D.-H.; Xue, S.-F.; Zhu, Q.-J.; Tao, Z. Structure of Supramolecular Assemblies Formed by α,δ-Tetramethylcucurbit[6]uril and 4-Nitrophenol. Molecules 2008, 13, 2814-2822. https://doi.org/10.3390/molecules13112814
Zheng L-M, Zhang Y-Q, Zeng J-P, Qiu Y, Yu D-H, Xue S-F, Zhu Q-J, Tao Z. Structure of Supramolecular Assemblies Formed by α,δ-Tetramethylcucurbit[6]uril and 4-Nitrophenol. Molecules. 2008; 13(11):2814-2822. https://doi.org/10.3390/molecules13112814
Chicago/Turabian StyleZheng, Li-Mei, Yun-Qian Zhang, Jin-Ping Zeng, Yan Qiu, Da-Hai Yu, Sai-Feng Xue, Qian-Jiang Zhu, and Zhu Tao. 2008. "Structure of Supramolecular Assemblies Formed by α,δ-Tetramethylcucurbit[6]uril and 4-Nitrophenol" Molecules 13, no. 11: 2814-2822. https://doi.org/10.3390/molecules13112814
APA StyleZheng, L. -M., Zhang, Y. -Q., Zeng, J. -P., Qiu, Y., Yu, D. -H., Xue, S. -F., Zhu, Q. -J., & Tao, Z. (2008). Structure of Supramolecular Assemblies Formed by α,δ-Tetramethylcucurbit[6]uril and 4-Nitrophenol. Molecules, 13(11), 2814-2822. https://doi.org/10.3390/molecules13112814