2. Results and Discussion
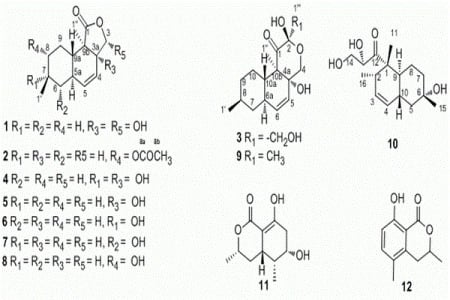
We obtained oblongolide C1 (
1) as white needles and determined it to have the molecular formula C
14H
20O
4 by HR-FT-MS. The
13C-NMR, DEPT and HSQC spectra of compound
1 showed 14 carbon signals: two methyl groups, three methylene groups, three methine groups, one hemiacetal methine (
δC 100.6), a disubstituted olefin (
δC 137.8 and 124.2), an oxygenated quaternary carbon (
δC 78.7), a lactone carbonyl (
δC 176.6), and a quaternary carbon. The
1H-
1H COSY correlations between H-4 and H-5, H-5 and H-5a, H-5a and H-6, H-5a and H-9a, H-6 and H-7, H-7 and H-1′, H-8 and H-7, H-8 and H-9, H-9a and H-9 established the structure of a 9-carbon moiety (
Figure 2, in green). Key HMBC correlations from H-1″ to C-1, C-3a, C-9a and C-9b, from H-5 to C-3a, from H-4 to C-9b, and from H-3 to C-3a established the planar structure of
1. The relative configuration of
1 was deduced on the basis of NOESY spectroscopic data. The NOE correlations between H-7 and H-5a and between H-5a and H-1″ established the α-orientations of H-5a, H-7 and H-1″. NOESY cross-peaks from H-3 to H-9a and from H-9a to H-1′ indicated the β-orientations of H-3, H-9a and H-1′. A comparison of the
1H and
13C-NMR spectra of
1 with that of oblongolide C indicated that
1 was the 3α-hydroxy derivative of oblongolide C [
4]. Therefore, we determined the structure of
1 to be 3α-hydroxyoblongolide C and it was named as oblongolide C1 for consistency with the literature [
4].
Figure 2.
Key HMBC and NOE Correlations of compound 1.
Figure 2.
Key HMBC and NOE Correlations of compound 1.
Oblongolide P1 (
2) was isolated as a white powder. The molecular formula C
16H
22O
4 was deduced from HR-FT-MS and
13C-NMR. NMR data of
2 were similar to those of
1, except that the hemiacetal methine [
δH 5.69 (1H, d,
J = 11.5 Hz) and
δC 100.6, CH-3], quaternary carbon (
δC 78.7, C-3a) and methylene [
δH 1.82 (1H, m),
δH 0.91 (1H, m) and
δC 34.6, CH-8] in
1 were replaced by oxymethylene [
δH 4.44 (1H, t,
J = 8.6 Hz),
δH 3.85 (1H, dd,
J = 10.9, 8.9 Hz) and
δC 70.1, CH
2-3], methine [
δH 2.78 (1H, m) and
δC 44.6, CH-3a], oxymethine [
δH 4.54 (1H, dt,
J = 10.8, 4.4 Hz) and
δC 77.1, CH-8], and there was an acetyl group in
2. Key HMBC correlations from H-8 to C-8a, C-1′ and C-9a, from H-1′ to C-6, C-7 and C-8, from H-1″ to C-1, C-3a, C-9a and C-9b indicated the planar structure of
2. We determined the relative configuration of
2 by analysis of the NOESY spectrum. The NOE correlations between H-8 and H-1′, between H-8 and H-9a, between H-8 and H-9β, and between H-1′ and H-6β established the β-orientations of H-1′, H-8 and H-9a. The NOE correlations between H-3a and H-1″, between H-3α and H-3a and between H-1″ and H-5a indicated the α-orientations of H-1″, H-3a and H-5a. A comparison of the
1H- and
13C-NMR data of
2 with those of oblongolide P [
3] revealed that these two compounds had similar structures, except that an acetyl group was attached to the C-8 hydroxyl group in
2. Therefore, we determined
2 to be 8-acetylobolngolide P and named it oblongolide P1.
Table 1.
1H- and 13C-NMR spectroscopic data of compounds 1 and 2 (1 and 2 at 600 MHz, CDCl3, chemical shift values are in ppm relative to TMS; multiplicity and J values (in Hz) are presented in parentheses.
Table 1.
1H- and 13C-NMR spectroscopic data of compounds 1 and 2 (1 and 2 at 600 MHz, CDCl3, chemical shift values are in ppm relative to TMS; multiplicity and J values (in Hz) are presented in parentheses.
| No. | 1 | 2 |
|---|
| δH | δC | δH | δC |
|---|
| 1 | | 176.6 | | 179.4 |
| 3α | | | 4.44 (t, 8.6) | 70.1 |
| 3β | 5.69 (d, 11.5) | 100.6 | 3.85 (dd, 8.9, 10.9) | 77.1 |
| 3a | | 78.7 | 2.78 (m) | 44.6 |
| 4 | 5.53 (dd, 10.2, 2.8) | 124.2 | 5.62 (dd, 12.8, 2.5) | 122.2 |
| 5 | 5.79 (d, 9.9) | 137.8 | 5.65 (d, 12.8) | 133.0 |
| 5a | 2.03 (m) | 36.3 | 1.97 (m) | 35.4 |
| 6α | 0.84 (q, 12.4) | 41.0 | 1.00 (m) | 39.0 |
| 6β | 1.90 (m) | 41.0 | 1.94 (m) | 39.0 |
| 7 | 1.50 (m) | 32.7 | 1.68 (m) | 37.4 |
| 8α | 0.91 (m) | 34.6 | | |
| 8β | 1.82 (m) | 34.6 | 4.54 (dt, 4.4, 10.8) | 77.1 |
| 9α | 1.34 (dd, 12.4, 3.1) | 25.6 | 1.38 (q, 12.2) | 31.1 |
| 9β | 1.79 (m) | 25.6 | 2.10 (m) | 31.1 |
| 9a | 1.49 (m) | 44.1 | 1.50 (m) | 37.3 |
| 9b | | 51.5 | | 42.8 |
| 1′ | 0.93 (d, 6.6) | 22.2 | 0.94 (d, 6.5) | 18.0 |
| 1″ | 1.18 (s) | 9.5 | 1.16 (s) | 16.1 |
| 8a | | | | 170.5 |
| 8b | | | 2.06 (s) | 21.1 |
Oblongolide X1 (
3) was obtained as white oil. Its molecular formula, C
16H
24O
5, was deduced on the basis of HR-FT-MS and
13C-NMR data. A comparison of the NMR data of
3 with those of known compound oblongolide X [
7] indicated that
3 was a hydroxy-derivative of the latter. The HMBC correlations from H-1″′ to C-1 and C-2 located the hydroxyl substitution at C-2. The NOE correlations between H-10a and H-1′, between H-6a and H-8 and between H-6a and H-1″ determined the relative configuration of
3. Therefore, we determined 3 to be 1″′-hydroxyoblongolide X and named it oblongolide X1.
Table 2.
1H- and 13C-NMR spectroscopic data of compound 3 (3 at 600 MHz, CDCl3, chemical shift values are in ppm relative to TMS; multiplicity and J values (in Hz) are presented in parentheses.
Table 2.
1H- and 13C-NMR spectroscopic data of compound 3 (3 at 600 MHz, CDCl3, chemical shift values are in ppm relative to TMS; multiplicity and J values (in Hz) are presented in parentheses.
| No. | 3 |
|---|
| δH | δC |
|---|
| 1 | - | 207.0 |
| 2 | - | 94.9 |
| 4α | 3.57 (d, 12.4) | 66.1 |
| 4β | 4.63 (d, 12.4) | 66.1 |
| 4a | | 78.2 |
| 5 | 5.36 (dd, 10.1, 2.8) | 126.9 |
| 6 | 5.68 (dd, 10.1, 1.6) | 136.2 |
| 6a | 1.93 (m) | 37.9 |
| 7α | 1.86 (m) | 41.1 |
| 7β | 0.89 (m) | 41.1 |
| 8 | 1.49 (m) | 33.0 |
| 9α | 1.77 (m) | 34.8 |
| 9β | 1.03 (m) | 34.8 |
| 10α | 1.26 (m) | 26.8 |
| 10β | 1.23 (m) | 26.8 |
| 10a | 2.33 (ddd, 11.5, 10.6 3.0) | 43.8 |
| 10b | | 55.4 |
| 1′ | 0.93 (d, 6.5) | 22.3 |
| 1‴ | 1.09 (s) | 10.4 |
| 1‴ | 3.61 (d, 11.9) | 65.2 |
| 1‴ β | 3.95 (d, 11.9) | 65.2 |
Compound
10 had the molecular formula C
16H
26O
4, as established by HR-FT-MS and
13C-NMR spectra.
1H- and
13C-NMR data of
10 were similar to those of phomodiol [
8], except that the methine signal [
δH 1.46 (1H, m), CH-6] was replaced by a quaternary carbon (
δC 70.0, C-6). Key HMBC correlations from H-15 to C-5, C-6 and C-7, from H-11 to C-1, C-2, C-9 and C-12, from H-16 to C-1, C-2 and C-3, from H-4 to C-2, C-5 and C-9 and from H-10 to C-3, C-6 and C-8 indicated the planar structure of
10. The relative configuration of
10 was deduced on the basis of NOESY spectroscopic data. The NOE correlations between H-10 and H-11, between H-2 and H-11, between H-13 and H-11, between H-15 and H-10 and between H-9 and H-16 indicated β-orientation of the hydroxyl group (6-OH) and the α-orientation of the side chain attached to C-1. Therefore, the structure of
10 was determined. We named it 6-hydroxyphomodiol [
8].
Table 3.
1H- and 13C-NMR spectroscopic data of compound 10 (600 MHz, in CDCl3, chemical shift values are in ppm relative to TMS; multiplicity and J values (in Hz) are presented in parentheses.
Table 3.
1H- and 13C-NMR spectroscopic data of compound 10 (600 MHz, in CDCl3, chemical shift values are in ppm relative to TMS; multiplicity and J values (in Hz) are presented in parentheses.
| No. | 10 |
|---|
| δH | δC |
|---|
| 1 | – | 5.15 |
| 2 | 2.18 (m) | 39.5 |
| 3 | 5.58 (ddd, 9.9, 4.9, 2.6) | 130.2 |
| 4 | 5.36 (d, 10.0) | 129.0 |
| 5α | 1.28 (m) | 45.5 |
| 5β | 1.75 (m) | 45.5 |
| 6 | – | 70.0 |
| 7α | 1.56 (dt, 13.6, 4.4) | 39.4 |
| 7β | 1.69 (dd, 14.1, 3.0) | 39.4 |
| 8α | 1.09 (brs) | 22.8 |
| 8β | 1.32 (m) | 22.8 |
| 9 | 1.79 (m) | 40.5 |
| 10 | 2.22 (m) | 33.0 |
| 11 | 1.35 (s) | 16.7 |
| 12 | – | 214.0 |
| 13 | 4.52 (brs) | 75.7 |
| 14 | 4.03 (dd, 11.8, 3.6), 3.79 (dd, 11.7, 4.7) | 63.3 |
| 15 | 1.27 (s) | 31.6 |
| 16 | 0.84 (d, 7.0) | 18.7 |
Besides the nine oblongolides, including three new ones, we isolated two more polyketides. We determined
11 to be (3
R,4a
R,5
S,6
R)-6-hydroxy-5-methylramulosin (
11) [
5] by a comparison of NMR data. This compound was previously isolated from a marine-derived fungus which was derived from the green alga
Codium fragile [
5]. The spectroscopic data of
12 were identical to those of the known compound (3
R)-5-methylmellein, first isolated as the main phytotoxic metabolite of
Fusicoccum amygdale [
6].
Cytotoxicity
The results of cytotoxic tests of compounds
1-
12 are shown in
Table 4. They exhibited no significant activity against the three tested cancer cell lines.
Table 4.
Biological Activities of Compounds 1-12.
Table 4.
Biological Activities of Compounds 1-12.
| Compound | Inhibitory rate(%) |
|---|
| HeLa | A549 | HepG2 |
|---|
| Oblongolide C1 (1) | - | - | 18.01 ± 0.86 |
| Oblongolide P1 (2) | - | - | 28.59 ± 1.04 |
| Oblongolide X1 (3) | - | - | 27.89 ± 1.2 |
| Oblongolide B (4) | - | - | - |
| Oblongolide C (5) | - | 14.92 ± 0.86 | - |
| Oblongolide D (6) | 22.9 ± 0.78 | 13.82 ± 1.01 | - |
| Oblongolide O (7) | - | - | - |
| Oblongolide P (8) | - | - | - |
| Oblongolide U (9) | - | 18.76 ± 0.56 | 16.89 ± 1.01 |
| 6-Hydroxyphomodiol (10) | - | - | 23.86 ± 1.21 |
| (3R,4aR,5S,6R)-6-Hydroxy-5-methylramulosin (11) | - | - | - |
| (3R)-5-Methylmellein (12) | - | - | - |
3. Experimental
3.1. General
Optical rotations were measured with a Perkin-Elmer 341 automatic polarimeter in methanol. IR spectra were recorded on a Nicolet AVATAR 330FT spectrometer. NMR spectra were taken on a Bruker Avance III-600 NMR spectrometer with TMS as an internal standard. HR-FT-MS data were acquired by using En Apex ultra 7.0 FT-MS. TLC was carried out using glass-precoated silica gel GF254 (Qingdao) and visualized under UV light or by spraying with vanillin (contains H2SO4) ethanol reagent. Sephadex LH-20 (40-70 µm, Amersham Pharmacia Biotech AB, Uppsala, Sweden), silica gel (200-300mesh, Qingdao Marine Chemical, Inc., Qingdao, China), and lichroprep reversed-phase RP-18 silica gel (40-63 µm, Merck, Darmstadt, Germany) were used for column chromatography (CC).
3.2. Fungal Material
The fungus (XZ-01) was isolated from current-year twigs (8-12 × 1-2 cm, length × diameter) of Camptotheca acuminate collected from the Jiangshi Natural Reserve, Shaowu, Fujian, China. It was identified as a non-sporulating fungus by traditional morphology. A BLAST search result showed that the internal transcribed spaces (ITS) sequence of XZ-01 was highly homologous (98% percent similarity) to that of a Phomopsis species (BCC 9789 [GU086404]), indicating that XZ-01 belongs to this genus.
3.3. Fermentation and Extraction
XZ-01 was cultivated on potato dextrose agar at 28 °C. The agar blocks were chopped and transferred into Erlenmeyer flasks (10 × 3 L), each containing 1 L of potato dextrose broth (PDB), and then fermented at 28 °C on a rotary shaker (150 rpm) for 7d. The culture was filtered to separate broth and mycelia. The culture broth was extracted with EtOAc (6 × 10 L) for six times. The combined organic layer was concentrated under vacuum to afford 3.2 g of residue.
3.4. Isolation and Spectral Data
The crude extract was separated into fifteen fractions (1-15) by column chromatography on RP-18 silica gel, eluted by methanol/H2O (0:100, 30:70, 50:50, 70:30, and 100:0). Fraction 3 (100 mg) was subjected to silica gel CC (step gradient, elution with 0-10% MeOH in CHCl3) to afford eleven fractions (3-1-3-11). Fractions 3-11 (4.9 mg) were further separated by silica gel CC (step gradient, elution with 22.2-33.3% EtOAc in hexane) to yield 4 (2.3 mg). Fraction 5 (92.1 mg) was separated by Sephadex LH-20 (elution with 100% methanol) to give three subfractions (fraction 5-1–5-3). Fraction 5-2 (23.6 mg) was purified by silica gel CC (step gradient, 7.7-50% EtOAc in hexane) to produce fraction 5-2-1. Fraction 5-2-1 (3.7 mg) was separated by silica gel (eluted with 50% CHCl3 in petroleum ether) to afford 11 (2mg). Fraction 6 (225.8 mg) was fractionated by Sephadex LH-20 CC (elution with 100% MeOH) to provide nine fractions (6-1–6-9). Fraction 6-1 (28.8 mg) was further purified by silica gel CC (step gradient, 0-17% MeOH in CHCl3) to furnish 6 (11.5 mg), 8 (2.6 mg) and 10 (6.4 mg). Fraction 7 (247.1 mg) was subjected to Sephadex LH-20 CC (elution with 100% MeOH) to give 5 fractions (7-1–7-5). Fraction 7-4 (36.1 mg) was purified by silica gel CC (elution with CHCl3) to yield 7 (3.1 mg). Fraction 10 (109 mg) was fractionated by Sephadex LH-20 CC (elution with 100% MeOH) to provide two fractions (10-1–10-2). Fraction 10-1 (72 mg) was further purified by silica gel CC (step gradient, elution with 0-10% MeOH in CHCl3) to give two subfractions (10-1-1 and 10-1-2). Fraction 10-1-2 (11.7 mg) was separated by silica gel CC (elution with 100% CHCl3) to yield 3 (3.8 mg). Fraction 11 (318.3 mg) was separated by Sephadex LH-20 (elution with 100% MeOH) to provide five fraction (11-1–11-5). Fraction 11-5 (23.9 mg) was further purified by silica gel CC (elution with 10% CHCl3 in petroleum ether) to afford 12 (22.8 mg). Fraction 11-3 (99 mg) was separated by silica gel CC (step gradient, elution with 0-10% MeOH in CHCl3) to give 5 (34 mg) and 9 (2.3 mg). Fraction 12 (117 mg) was fractionated by Sephadex LH-20 CC (elution with 100% MeOH) to provide three fractions (12-1–12-3). Fraction 12-1 (12.8 mg) was further separated by silica gel CC (elution with 33.3% CHCl3 in petroleum ether) to yield 2 (7.4 mg). Fraction 9 (232 mg) was separated by Sephadex LH-20 (elution with 100% MeOH) to give two fractions (9-1–9-2). Fraction 9-2 (38 mg) was purified by silica gel CC (step gradient, 0-12.3% MeOH in CHCl3) to yield 1 (5.7 mg).
Oblongolide C-1 (
1): White needles; [α]
D20: – 22.6 (c 0.0072, MeOH). IR (KBr) ν
max 2919, 2359, 1219, 772, 668 cm
–1.
1H- and
13C-NMR: see
Table 1; HR-FT-MS: m/z = 251.1281 [M − H]
- (calcd. for C
14H
19O
4, 251.1283, Temperature: 180, Resolution: 125,508).
Oblongolide O-1 (
2): White powder; [α]
D20: – 72.2(c 0.0025, MeOH). IR (KBr) ν
max 3344, 2922, 1588, 1383, 772 cm
–1.
1H- and
13C-NMR: see
Table 1; HR-FT-MS: m/z = 301.1418 [M + Na]
+ (calcd. for C
16H
22O
4Na, 301.1416, Temperature: 180, Resolution: 14,100).
Oblongolide X-1 (
3): White oil; [α]
D20: – 21.7(c 0.0056, MeOH). IR (KBr) ν
max 3422, 1583, 773, 685 cm
–1.
1H- and
13C-NMR: see
Table 2; HR-FT-MS: m/z = 295.1541 [M − H]
- (calcd. for C
16H
21O
5, 295.1545, Temperature: 180, Resolution: 106,466).
6-Hydroxyphomodiol (
10): Transparent oil; [α]
D20: + 43.3(c 0.002, MeOH). IR (KBr) ν
max 2365, 1223, 771 cm
–1.
1H- and
13C-NMR: see
Table 3; HR-FT-MS: m/z = 305.1736 [M + Na]
- (calcd. for C
16H
26NaO
4, 305.1729, Temperature: 180, Resolution: 36,000).
3.5. Biological Assay
Cancer cell lines were derived from the cell bank of The Chinese Academy of Sciences. Cells were seeded at a density of 5 × 103/100 µL medium in 96-well microtiter plate and treated with the compounds at the concentration of 20 µg/mL. Viable cells were incubated with MTT (5 mg/mL) for 4 h and formazan precipitate was dissolved in 100 µL DMSO and the absorbance at 490 nm was measured by Multimode Detector DTX880 (Beckman Coulter).