Accumulation of Unusual Gangliosides GQ3 and GP3 in Breast Cancer Cells Expressing the GD3 Synthase
Abstract
:Abbreviations:
| BSA: | Bovine Serum Albumin |
| Cer: | ceramide |
| DMB: | 1,2-diamino-4,5-methylenedioxybenzene |
| DMEM: | Dulbecco’s Modified Eagle’s Medium |
| EDTA: | Ethylenediaminetetraacetic Acid |
| DMSO: | Dimethyl Sulfoxide |
| FBS: | Fetal Bovine Serum |
| FITC: | Fluorescein Isothiocyanate |
| FL-HPLC: | Fluorescence Detection High Performance Liquid Chromatography |
| GD3S: | GD3 synthase |
| GSL: | glycosphingolipid |
| HPRT: | Hypoxanthine PhosphoRibosylTransferase |
| HRP: | horseradish peroxidase |
| LacCer: | Lactosylceramide |
| mAb: | monoclonal Antibody |
| MALDI-TOF: | matrix assisted laser desorption-ionization time-of-flight |
| MS: | Mass Spectrometry |
| PBS: | Phosphate Buffered Saline |
| QPCR: | Quantitative real-time Polymerase Chain Reaction |
| SDS-PAGE: | Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis |
| ST3Gal V: | GM3 synthase |
| ST8Sia I: | GD3 synthase |
| ST8Sia V: | GT3 synthase; WT: Wild Type |
1. Introduction

| Gene | Common name | Main acceptor(s) | Accession # | Reference |
|---|---|---|---|---|
| UGCG | GlcCer synthase | Ceramide | NM_003358 | [18] |
| B4GALT6 | LacCer synthase | Glucosylceramide | NM_004775 | [19,20] |
| ST3GAL5 | GM3 synthase | Lactosylceramide | NM_003896 | [21] |
| ST8SIA1 | GD3 synthase | GM3, GD3 | NM_003034.2 | [11,12,13] |
| ST8SIA5 | GT3 synthase | GD3, GM1b, GD1a, GT1b | NM_013305 | [17] |
| B4GALNACT1 | GM2/GD2 synthase | GA3, GM3, GD3, GT3 | NM_001478.2 | [22,23,24] |
| B3GALT4 | GM1a/GD1b synthase | GA2, GM2, GD2, GT2 | NM_003782.3 | [23,25] |
| ST3GAL1 | ST3Gal I | Galβ1-3GalNAcβ1-4-R | NM_003033 | [26] |
| ST3GAL2 | ST3Gal II | Galβ1-3GalNAcβ1-4-R | NM_006927 | [27] |
| ST6GALNAC3 | ST6GalNAc III | Neu5Acα2-3Galβ1-3GalNAcβ1-4-R | NM_152996 | [28] |
| ST6GALNAC5 | ST6GalNAc V | Neu5Acα2-3Galβ1-3GalNAcβ1-4-R | NM_030965.1 | [29] |
| ST6GALNAC6 | ST6GalNAc VI | Neu5Acα2-3Galβ1-3GalNAcβ1-4-R | NM_013443.3 | [29] |
2. Results and Discussion
2.1. Analysis of ST8Sia I Expression by QPCR in Control and GD3S+ MCF-7 Clones

2.2. Flow Cytometry Analysis of Gangliosides Expression in MCF-7 GD3S+ Clones
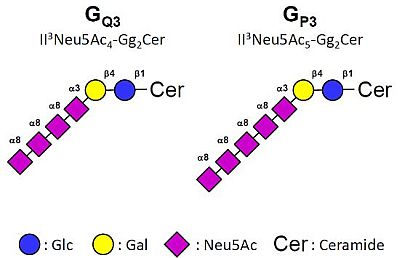
2.3. MS Analysis of Gangliosides in MCF-7 and GD3S+ Clones

 , Gal;
, Gal;  , Glc;
, Glc;  , GalNAc;
, GalNAc;  , Neu5Ac.
, Neu5Ac.
 , Gal;
, Gal;  , Glc;
, Glc;  , GalNAc;
, GalNAc;  , Neu5Ac.
, Neu5Ac.
 , Gal;
, Gal;  , Glc;
, Glc;  , GalNAc;
, GalNAc;  , Neu5Ac.
, Neu5Ac.
 , Gal;
, Gal;  , Glc;
, Glc;  , GalNAc;
, GalNAc;  , Neu5Ac.
, Neu5Ac.
2.4. Quantification of Polysialylation Associated with Gangliosides by HPLC

3. Experimental Section
3.1. Antibodies and Reagents
3.2. Cell Culture
3.3. Quantitative Real-Time-PCR (QPCR) Analysis of GD3 Synthase
3.4. Analysis of Cell Surface Ganglioside by Flow Cytometry
3.5. Extraction and Preparation of Glycolipids
3.6. Mass Spectrometry Analysis of GSL
3.7. Analysis of Oligo-Sialylated Sequences by HPLC
4. Conclusions
Acknowledgments
References
- Svennerholm, L. Ganglioside designation. Adv. Exp. Med. Biol. 1980, 125, 11–19. [Google Scholar]
- Zeng, G.; Yu, R.K. Cloning and transcriptional regulation of genes responsible for synthesis of gangliosides. Curr. Drug Targets 2008, 9, 317–324. [Google Scholar] [CrossRef]
- Yamamoto, A.; Haraguchi, M.; Yamashiro, S.; Fukumoto, S.; Furukawa, K.; Takamiya, K.; Atsuta, M.; Shiku, H.; Furukawa, K. Heterogeneity in the expression pattern of two ganglioside synthase genes during mouse brain development. J. Neurochem. 1996, 66, 26–34. [Google Scholar]
- Yu, R.K.; Macala, L.J.; Taki, T.; Weinfield, H.M.; Yu, F.S. Developmental changes in ganglioside composition and synthesis in embryonic rat brain. J. Neurochem. 1988, 50, 1825–1829. [Google Scholar]
- Yamashita, T.; Wada, R.; Sasaki, T.; Deng, C.; Bierfreund, U.; Sandhoff, K.; Proia, R.L. A vital role for glycosphingolipid synthesis during development and differentiation. Proc. Natl. Acad. Sci. USA 1999, 96, 9142–9147. [Google Scholar]
- Nakayama, J.; Fukuda, M.N.; Hirabayashi, Y.; Kanamori, A.; Sasaki, K.; Nishi, T.; Fukuda, M. Expression cloning of a human GT3 synthase. GD3 and GT3 are synthesized by a single enzyme. J. Biol. Chem. 1996, 271, 3684–3691. [Google Scholar]
- Furukawa, K.; Hamamura, K.; Aixinjueluo, W.; Furukawa, K. Biosignals modulated by tumor-associated carbohydrate antigens novel targets for cancer therapy. Ann. NY Acad. Sci. 2006, 1086, 185–198. [Google Scholar] [CrossRef]
- Oblinger, J.L.; Pearl, D.K.; Boardman, C.L.; Saqr, H.; Prior, T.W.; Scheithauer, B.W.; Jenkins, R.B.; Burger, P.C.; Yates, A.J. Diagnostic and prognostic value of glycosyltransferase mRNA in glioblastoma multiforme patients. Neuropathol. Appl. Neurobiol. 2006, 32, 410–418. [Google Scholar] [CrossRef]
- Ruan, S.; Lloyd, K.O. Glycosylation pathways in the biosynthesis of gangliosides in melanoma and neuroblastoma cells relative glycosyltransferase levels determine ganglioside patterns. Cancer Res. 1992, 52, 5725–5731. [Google Scholar]
- Ruckhäberle, E.; Rody, A.; Engels, K.; Gaetje, R.; von Minckwitz, G.; Schiffmann, S.; Grösch, S.; Geisslinger, G.; Holtrich, U.; Karn, T.; Kaufmann, M. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res. Treat. 2008, 112, 41–52. [Google Scholar] [CrossRef]
- Nara, K.; Watanabe, Y.; Maruyama, K.; Kasahara, K.; Nagai, Y.; Sanai, Y. Expression cloning of a CMP-NeuAcNeuAc alpha 2-3Gal beta 1-4Glc beta 1-1'Cer alpha 2,8-sialyltransferase (GD3 synthase) from human melanoma cells. Proc. Natl. Acad. Sci. USA 1994, 91, 7952–7956. [Google Scholar] [CrossRef]
- Sasaki, K.; Kurata, K.; Kojima, N.; Kurosawa, N.; Ohta, S.; Hanai, N.; Tsuji, S.; Nishi, T. Expression cloning of a GM3-specific alpha-2,8-sialyltransferase (GD3 synthase). J. Biol. Chem. 1994, 269, 15950–15956. [Google Scholar]
- Haraguchi, M.; Yamashiro, S.; Yamamoto, A.; Furukawa, K.; Takamiya, K.; Lloyd, K.O.; Shiku, H.; Furukawa, K. Isolation of GD3 synthase gene by expression cloning of GM3 alpha-2,8-sialyltransferase cDNA using anti-GD2 monoclonal antibody. Proc. Natl. Acad. Sci. USA 1994, 91, 10455–10459. [Google Scholar]
- Furukawa, K.; Horie, M.; Okutomi, K.; Sugano, S.; Furukawa, K. Isolation and functional analysis of the melanoma specific promoter region of human GD3 synthase gene. Biochim. Biophys. Acta 2003, 1627, 71–78. [Google Scholar] [CrossRef]
- Harduin-Lepers, A.; Vallejo-Ruiz, V.; Krzewinski-Recchi, M.A.; Samyn-Petit, B.; Julien, S.; Delannoy, P. The human sialyltransferase family. Biochimie 2001, 83, 727–737. [Google Scholar]
- Nara, K.; Watanabe, Y.; Kawashima, I.; Tai, T.; Nagai, Y.; Sanai, Y. Acceptor substrate specificity of a cloned GD3 synthase that catalyzes the biosynthesis of both GD3 and GD1c/GT1a/GQ1b. Eur. J. Biochem. 1996, 238, 647–652. [Google Scholar]
- Kim, Y.J.; Kim, K.S.; Do, S.; Kim, C.H.; Kim, S.K.; Lee, Y.C. Molecular cloning and expression of human alpha2,8-sialyltransferase (hST8Sia V). Biochem. Biophys. Res. Commun. 1997, 235, 327–330. [Google Scholar]
- Ichikawa, S.; Sakiyama, H.; Suzuki, G.; Hidari, K.I.; Hirabayashi, Y. Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc. Natl. Acad. Sci. USA 1996, 93, 4638–4643. [Google Scholar]
- Nomura, T.; Takizawa, M.; Aoki, J.; Arai, H.; Inoue, K.; Wakisaka, E.; Yoshizuka, N.; Imokawa, G.; Dohmae, N.; Takio, K.; et al. Purification, cDNA cloning, and expression of UDP-Gal: Glucosylceramide beta-1,4-galactosyltransferase from rat brain. J. Biol. Chem. 1998, 273, 13570–13577. [Google Scholar]
- Takizawa, M.; Nomura, T.; Wakisaka, E.; Yoshizuka, N.; Aoki, J.; Arai, H.; Inoue, K.; Hattori, M.; Matsuo, N. cDNA cloning and expression of human lactosylceramide synthase. Biochim. Biophys. Acta 1999, 1438, 301–304. [Google Scholar] [CrossRef]
- Ishii, A.; Ohta, M.; Watanabe, Y.; Matsuda, K.; Ishiyama, K.; Sakoe, K.; Nakamura, M.; Inokuchi, J.; Sanai, Y.; Saito, M. Expression cloning and functional characterization of human cDNA for ganglioside GM3 synthase. J. Biol. Chem. 1998, 273, 31652–31655. [Google Scholar]
- Nagata, Y.; Yamashiro, S.; Yodoi, J.; Lloyd, K.O.; Shiku, H.; Furukawa, K. Expression cloning of beta 1,4 N-acetylgalactosaminyltransferase cDNAs that determine the expression of GM2 and GD2 gangliosides. J. Biol. Chem. 1992, 267, 12082–12089. [Google Scholar]
- Iber, H.; Zacharias, C.; Sandhoff, K. The c-series gangliosides GT3, GT2 and GP1c are formed in rat liver Golgi by the same set of glycosyltransferases that catalyse the biosynthesis of asialo-, a- and b-series gangliosides. Glycobiology 1992, 2, 137–142. [Google Scholar] [CrossRef]
- Yamashiro, S.; Haraguchi, M.; Furukawa, K.; Takamiya, K.; Yamamoto, A.; Nagata, Y.; Lloyd, K.O.; Shiku, H.; Furukawa, K. Substrate specificity of beta 1,4-N-acetylgalactosaminyltransferase in vitro and in cDNA-transfected cells. GM2/GD2 synthase efficiently generates asialo-GM2 in certain cells. J. Biol. Chem 1995, 270, 6149–6155. [Google Scholar]
- Amado, M.; Almeida, R.; Carneiro, F.; Levery, S.B.; Holmes, E.H.; Nomoto, M.; Hollingsworth, M.A.; Hassan, H.; Schwientek, T.; Nielsen, P.A.; et al. A family of human beta3-galactosyltransferases. Characterization of four members of a UDP-galactose: Beta-N-acetyl-glucosamine/beta-N-acetyl-galactosamine beta-1,3-galactosyltransferase family. J. Biol. Chem 1998, 273, 12770–12778. [Google Scholar]
- Kitagawa, H.; Paulson, J.C. Differential expression of five sialyltransferase genes in human tissues. J. Biol. Chem. 1994, 269, 17872–17878. [Google Scholar]
- Giordanengo, V.; Bannwarth, S.; Laffont, C.; van Miegem, V.; Harduin-Lepers, A.; Delannoy, P.; Lefebvre, J.C. Cloning and expression of cDNA for a human Gal(beta1-3)GalNAc alpha2,3-sialyltransferase from the CEM T-cell line. Eur. J. Biochem. 1997, 247, 558–566. [Google Scholar]
- Tsuchida, A.; Ogiso, M.; Nakamura, Y.; Kiso, M.; Furukawa, K.; Furukawa, K. Molecular cloning and expression of human ST6GalNAc III: Restricted tissue distribution and substrate specificity. J. Biochem. 2005, 138, 237–243. [Google Scholar]
- Harduin-Lepers, A.; Mollicone, R.; Delannoy, P.; Oriol, R. The animal sialyltransferases and sialyltransferase-related genes: A phylogenetic approach. Glycobiology 2005, 15, 805–817. [Google Scholar] [CrossRef]
- Cazet, A.; Groux-Degroote, S.; Teylaert, B.; Kwon, K.M.; Lehoux, S.; Slomianny, C.; Kim, C.H.; Le Bourhis, X.; Delannoy, P. GD3 synthase overexpression enhances proliferation and migration of MDA-MB-231 breast cancer cells. Biol. Chem. 2009, 390, 601–609. [Google Scholar]
- Ruan, S.; Raj, B.K.; Lloyd, K.O. Relationship of glycosyltransferases and mRNA levels to ganglioside expression in neuroblastoma and melanoma cells. J. Neurochem. 1999, 72, 514–521. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Steenackers, A.; Cazet, A.; Bobowski, M.; Rombouts, Y.; Lefebvre, J.; Guérardel, Y.; Tulasne, D.; Le Bourhis, X.; Delannoy, P. Expression of GD3 synthase modifies ganglioside profile and increases migration of MCF-7 breast cancer cells. C.R. Chim. 2012, 15, 3–14. [Google Scholar]
- Domon, B.; Costello, C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjug. J. 1988, 5, 397–409. [Google Scholar] [CrossRef]
- Sato, C.; Inoue, S.; Matsuda, T.; Kitajima, K. Fluorescent-assisted detection of oligosialyl units in glycoconjugates. Anal. Biochem. 1999, 266, 102–109. [Google Scholar]
- Chang, L.-Y.; Harduin-Lepers, A.; Kitajima, K.; Sato, C.; Huang, C.-J.; Khoo, K.-H.; Guérardel, Y. Developmental regulation of oligosialylation in zebrafish. Glycoconjug. J. 2009, 26, 247–261. [Google Scholar]
- Moon, S.K.; Kim, H.M.; Lee, Y.C.; Kim, C.H. Disialoganglioside (GD3) synthase gene expression suppresses vascular smooth muscle cell responses via the inhibition of ERK1/2 phosphorylation, cell cycle progression, and matrix metalloproteinase-9 expression. J. Biol. Chem. 2004, 279, 33063–33070. [Google Scholar]
- Zhang, X.; Ding, L.; Sandford, A.J. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol. Biol. 2005, 6, 4. [Google Scholar]
- Schnaar, R.L. Isolation of glycosphingolipids. Methods Enzymol. 1994, 230, 348–370. [Google Scholar]
- Ciucanu, I.; Kerek, F. Rapid and simultaneous methylation of fatty and hydroxy fatty acids for gas-liquid chromatographic analysis. J. Chromatogr. 1984, 284, 179–185. [Google Scholar]
- Harduin-Lepers, A.; Petit, D.; Mollicone, R.; Delannoy, P.; Petit, J.-M.; Oriol, R. Evolutionary history of the alpha2,8-sialyltransferase (ST8Sia) gene family: Tandem duplications in early deuterostomes explain most of the diversity found in the vertebrate ST8Sia genes. BMC Evol. Biol. 2008, 8, 258. [Google Scholar]
- Miyata, S.; Yamakawa, N.; Toriyama, M.; Sato, C.; Kitajima, K. Co-expression of two distinct polysialic acids, α2,8- and α2,9-linked polymers of N-acetylneuraminic acid, in distinct glycoproteins and glycolipids in sea urchin sperm. Glycobiology 2011, 21, 1596–1605. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Steenackers, A.; Vanbeselaere, J.; Cazet, A.; Bobowski, M.; Rombouts, Y.; Colomb, F.; Bourhis, X.L.; Guérardel, Y.; Delannoy, P. Accumulation of Unusual Gangliosides GQ3 and GP3 in Breast Cancer Cells Expressing the GD3 Synthase. Molecules 2012, 17, 9559-9572. https://doi.org/10.3390/molecules17089559
Steenackers A, Vanbeselaere J, Cazet A, Bobowski M, Rombouts Y, Colomb F, Bourhis XL, Guérardel Y, Delannoy P. Accumulation of Unusual Gangliosides GQ3 and GP3 in Breast Cancer Cells Expressing the GD3 Synthase. Molecules. 2012; 17(8):9559-9572. https://doi.org/10.3390/molecules17089559
Chicago/Turabian StyleSteenackers, Agata, Jorick Vanbeselaere, Aurélie Cazet, Marie Bobowski, Yoann Rombouts, Florent Colomb, Xuefen Le Bourhis, Yann Guérardel, and Philippe Delannoy. 2012. "Accumulation of Unusual Gangliosides GQ3 and GP3 in Breast Cancer Cells Expressing the GD3 Synthase" Molecules 17, no. 8: 9559-9572. https://doi.org/10.3390/molecules17089559
APA StyleSteenackers, A., Vanbeselaere, J., Cazet, A., Bobowski, M., Rombouts, Y., Colomb, F., Bourhis, X. L., Guérardel, Y., & Delannoy, P. (2012). Accumulation of Unusual Gangliosides GQ3 and GP3 in Breast Cancer Cells Expressing the GD3 Synthase. Molecules, 17(8), 9559-9572. https://doi.org/10.3390/molecules17089559