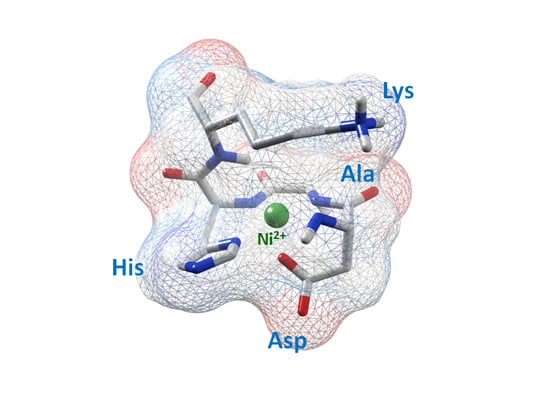
The Involvement of Amino Acid Side Chains in Shielding the Nickel Coordination Site: An NMR Study
Abstract
:1. Introduction

2. Results and Discussion
2.1. Albumin

2.2. Des-Angiotensiogen
| Peptide | Protein | Ref. | logK * | Coordination sphere | |
|---|---|---|---|---|---|
| Boc-AGGH | [18] | −30.02 | {NIm, 3N−} | ||
| Ac-ELAKHA-Am | Histone H2B | [19] | −28.87 | {NIm, 3N−} | |
| Ac-IQTAVRLLLPGELAKHAVSEGTKAVTKYTSSK-Am | Histone H2B | [20] | −28.83 | {NIm, 3N−} | |
| Ac-AKRHRK-Am | Histone H4 | [21] | −28.70 | {NIm, 3N−} | |
| Ac- SGRGKGGKGLGKGGAKRHRKVL -Am | Histone H4 | [22] | −28.67 | {NIm, 3N−} | |
| Ac-TESHHK-Am | Histone H2A | [23] | −28.58 | {NIm, 3N−} | |
| Ac-TEAHHK-Am | Histone H2A * | [23] | −28.41 | {NIm, 3N−} | |
| Ac-TESHAK-Am | Histone H2A * | [23] | −28.23 | {NIm, 3N−} | |
| Ac-AK(Ac)RHRK(Ac)V-Am | Histone H4 | [22] | −28.20 | {NIm, 3N−} | |
| Ac-TESAHK-Am | Histone H2A * | [23] | −28.18 | {NIm, 3N−} | |
| Ac-TRSRSHTSEGTRSR-Am | Cap43 | [24] | −28.16 | {NIm, 3N−} | |
| Ac-TYTEHA-Am | Histone H4 | [25] | −27.92 | {NIm, 3N−} | |
| Ac-TASHHK-Am | Histone H2A * | [23] | −27.26 | {NIm, 3N−} | |
| NH2-GGHistamine | [26] | −22.65 | {Nim, 2N−, NH2} | ||
| NH2-GGH | [15] | −21.81 | {Nim, 2N−, NH2} | ||
| NH2-SAHK-Am | Histone H2A * | [27] | −21.80 | {Nim, 2N−, NH2} | |
| NH2-VIHN | Des-Angiotensinogen | [2] | −19.75 | {Nim, 2N−, NH2} | |
| NH2-DAHK-Am | Albumin | [15] | −19.48 | {Nim, 2N−, NH2, O-} | |
| NH2-RTHGQSHYRRRHCSR-Am | Protamine HP2 | [28] | −19.29 | {Nim, 2N−, NH2} | |
| NH2-RTHGQ-Am | Protamine HP2 | [28] | −19.23 | {Nim, 2N−, NH2} | |
| NH2-SHHK-Am | Histone H2A | [27] | −19.14 | {Nim, 2N−, NH2} | |
2.3. Histones
2.3.1. Histone H4
2.3.2. Histone H2A
2.3.3. Histone H2B
2.4. Prion Proteins
2.5. Protamine
2.6. Cap43 Protein
3. Discussion
| XY-His-Z sequence | Protein | Ref. | X | Y | His | Z | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hα | Hβ2 | Hβ3 | Hα | Hβ2 | Hβ3 | Hα | Hβ2 | Hβ3 | Hδ2 | Hε1 | Hα | Hβ2 | Hβ3 | |||
| DEKHE | YPK9 | * | −0.23 | 0.70 | 0.56 | −0.59 | 0.57 | −0.17 | −1.33 | −0.12 | −0.16 | −0.06 | 0.01 | 0.01 | 0.02 | 0.00 |
| LAKHA | Histone H2B | [20] | −0.60 | −0.18 | −0.18 | −1.39 | 0.18 | 0.06 | −1.31 | −0.21 | −0.18 | −0.05 | −0.12 | −0.25 | −0.06 | −0.06 |
| RLAHY | Histone H2B | [39] | −0.18 | 0.49 | 0.61 | −0.98 | −0.04 | −0.04 | −1.15 | −0.14 | −0.24 | 0.13 | −0.29 | 0.09 | 0.16 | 0.11 |
| MKHM | Prion Protein | [57] | −0.41 | 0.41 | 0.34 | −0.56 | 0.13 | 0.04 | −1.12 | −0.21 | 0.03 | 0.00 | −0.20 | 0.10 | 0.23 | 0.13 |
| SRSHT | Cap43 | [52] | −0.31 | 0.41 | 0.35 | −0.32 | −0.15 | −0.15 | −1.09 | −0.17 | −0.17 | −0.02 | −0.18 | 0.04 | 0.16 | |
| AKRHR | Histone H4 | [35] | −0.26 | 0.28 | 0.39 | −0.67 | 0.36 | 0.55 | −1.03 | −0.17 | −0.23 | −0.04 | −0.13 | 0.05 | 0.16 | 0.03 |
| SAHK | Histone H2A | [27] | 0.51 | −0.79 | −0.62 | −0.61 | -0.06 | −0.78 | 0.34 | −0.04 | −0.19 | −0.01 | −0.07 | |||
| SHHK | Histone H2A | [27] | 0.44 | −0.69 | −0.45 | −0.72 | -0.82 | −0.64 | 0.42 | −0.04 | −0.25 | −0.03 | −0.07 | |||
| VIHN | Angiotensin | [2] | 0.17 | 0.31 | −0.36 | 0.05 | −0.57 | −0.26 | −0.01 | −0.09 | −0.28 | −0.22 | −0.06 | −0.29 | ||
| DAHK | Albumin | [6] | −0.24 | −0.06 | −0.06 | −0.56 | −0.05 | −0.05 | −0.47 | −0.15 | −0.11 | −0.05 | −0.24 | |||
| RTHG | Protamine | [43] | 0.59 | 0.03 | 0.18 | −0.20 | −0.37 | −0.45 | 0.01 | −0.20 | −0.06 | −0.27 | −0.10 | |||
| GTHS | Prion | [41] | −0.65 | −0.34 | −0.38 | −0.44 | −0.01 | −0.08 | −0.09 | −0.22 | −0.28 | −0.36 |
| XY-His-Z sequence | Protein | Ref. | X | Y | His | Z | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cα | Cβ | Cα | Cβ | Cα | Cβ | Cδ2 | Cε1 | Cα | Cβ | |||||||
| DEKHE | YPK9 | * | 10.77 | 6.87 | 6.21 | −1.54 | 3.17 | 1.45 | −3.70 | 1.51 | −0.11 | −0.03 | ||||
| SRSHT | Cap43 | [52] | 11.07 | −3.55 | 6.67 | 1.11 | 1.26 | 1.36 | −3.23 | 1.02 | −0.28 | −0.09 | ||||
| RLAHY | Histone H2B | [39] | 10.9 | NA | 5.9 | 1.7 | 1.9 | NA | NA | NA | −1.6 | NA | ||||
| LAKHA | Histone H2B | [20] | 10.1 | 0.73 | 5.31 | NA | 3.09 | 1.45 | NA | NA | 3.98 | 3.58 | ||||
| DAHK | Albumin | [6] | NA | NA | NA | NA | NA | NA | −2.59 | 1.1 | NA | NA | ||||
| GTHS | Prion | [41] | 5.5 | −1.2 | 7 | −1.2 | −3 | 0.7 | −2 | 0 | 0 | 0 |

4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Bal, W.; Kozlowski, H.; Kupryszewski, G.; Mackiewicz, Z.; Pettit, L.; Robbins, R. Complexes of Cu(II) with Asn-Ser-Phe-Arg-Tyr-NH2; an example of metal ion-promoted conformational organization which results in exceptionally high complex stability. J. Inorg. Biochem. 1993, 52, 79–87. [Google Scholar] [CrossRef]
- Bal, W.; Chmurny, G.N.; Hilton, B.D.; Sadler, P.J.; Tucker, A. Axial hydrophobic fence in highly stable Ni(II) complex of des-angiotensinogen N-terminal peptide. J. Am. Chem. Soc. 1996, 118, 4727–4728. [Google Scholar] [CrossRef]
- Handbook of Metalloproteins; Bertini, I.; Sigel, A.; Sigel, H. (Eds.) Marcel Dekker Inc.: New York, NY, USA, 2001.
- Handbook of Metalloproteins; Messerschmidt, A.; Huber, R.; Poulos, T.; Wieghardt, K. (Eds.) John Wiley & Sons, Ltd.: Chichester, UK, 2001; Volume 2.
- Beck, M.T. Critical evaluation of equilibrium constants in solution. Stability constants of metal complexes. Pure Appl. Chem. 1977, 49, 127–136. [Google Scholar] [CrossRef]
- Laussac, J.P.; Sarkar, B. Characterization of the copper(II)- and nickel(II)-transport site of human serum albumin. Studies of copper(II) and nickel(II) binding to peptide 1-24 of human serum albumin by 13C and 1H NMR spectroscopy. Biochemistry 1984, 23, 2832–2938. [Google Scholar] [CrossRef]
- Sadler, P.J.; Tucker, A.; Viles, J.H. Involvement of a lysine residue in the N-terminal Ni2+ and Cu2+ binding site of serum albumins. Comparison with Co2+, Cd2+ and Al3+. Eur. J. Biochem. 1994, 220, 193–200. [Google Scholar] [CrossRef]
- Sadler, P.J.; Tucker, A. 1H n.m.r. studies of serum albumin: assignment of resonances for N-terminal amino acids. Biochem Soc Trans 1990, 18, 923–924. [Google Scholar]
- Sadler, P.J.; Tucker, A. Proton NMR studies of bovine serum albumin. Assignment of spin systems. Eur. J. Biochem. 1992, 205, 631–643. [Google Scholar] [CrossRef]
- Froimowitz, M. HyperChem: A software package for computational chemistry and molecular modeling. Biotechniques 1993, 14, 1010–1013. [Google Scholar]
- Guntert, P.; Mumenthaler, C.; Wuthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Perkins, C.M.; Rose, N.J.; Weinstein, B.; Stenkamp, R.E.; Jensen, L.H.; Pickart, L. The structure of a copper complex of the growth factor glycyl-l-histidyl-l-lysine at 1.1 angstrom resolution. Inorg. Chem. Acta 1984, 82, 93–99. [Google Scholar] [CrossRef]
- Perkins, S.J. Application of ring current calculations to the proton NMR of protein and transfer RNA. Biol. Magn. Reson. 1982, 4, 193–336. [Google Scholar] [CrossRef]
- Sokolowska, M.; Krezel, A.; Dyba, M.; Szewczuk, Z.; Bal, W. Short peptides are not reliable models of thermodynamic and kinetic properties of the N-terminal metal binding site in serum albumin. Eur. J. Biochem. 2002, 269, 1323–1331. [Google Scholar] [CrossRef]
- Camerman, N.; Camerman, A.; Sarkar, B. Molecular design to mimic the copper(II) transport site of human albumin. The crystal and molecular structure of copper(II) - glycylglycyl-L-histidine-Nmethyl amide monoaquo complex. Can. J. Chem. 1976, 54, 1309–1316. [Google Scholar] [CrossRef]
- Pettit, G.; Pettit, L.D. IUPAC Stability Constants Database; IUPAC and Academic Software: Leeds, UK, 1993. [Google Scholar]
- Bal, W.; Kozlowski, H.; Pettit, L.D.; Robbins, R. Competition between the terminal amino and imidazole nitrogen donors for co−ordination to Ni(II) ions in oligopeptides. Inorg. Chim. Acta 1995, 231, 7–12. [Google Scholar] [CrossRef]
- Karavelas, T.; Mylonas, M.; Malandrinos, G.; Plakatouras, J.C.; Hadjiliadis, N.; Mlynarz, P.; Kozlowski, H. Coordination properties of Cu(II) and Ni(II) ions towards the C-terminal peptide fragment -ELAKHA- of histone H2B. J. Inorg. Biochem. 2005, 99, 606–615. [Google Scholar] [CrossRef]
- Nunes, A.M.; Zavitsanos, K.; Del Conte, R.; Malandrinos, G.; Hadjiliadis, N. The possible role of 94-125 peptide fragment of histone H2B in nickel-induced carcinogenesis. Inorg. Chem. 2010, 49, 5658–5668. [Google Scholar] [CrossRef]
- Zoroddu, M.A.; Kowalik-Jankowska, T.; Kozlowski, H.; Molinari, H.; Salnikow, K.; Broday, L.; Costa, M. Interaction of Ni(II) and Cu(II) with a metal binding sequence of histone H4: AKRHRK, a model of the H4 tail. Biochim. Biophys. Acta 2000, 1475, 163–168. [Google Scholar] [CrossRef]
- Zoroddu, M.A.; Peana, M.; Kowalik-Jankowska, T.; Kozlowski, H.; Costa, M. The binding of Ni(II) and Cu(II) with the N-terminal tail of the histone H4. J. Chem. Soc. Dalton. Trans. 2002, 3, 458–465. [Google Scholar]
- Mylonas, M.; Krezel, A.; Plakatouras, J.C.; Hadjiliadis, N.; Bal, W. The binding of Ni(II) ions to terminally blocked hexapeptides derived from the metal binding -ESHH- motif of histone H2A. J. Chem. Soc. Dalton. Trans. 2002, 4296–4306. [Google Scholar]
- Zoroddu, M.A.; Kowalik-Jankowska, T.; Kozlowski, H.; Salnikow, K.; Costa, M. Ni(II) and Cu(II) binding with a 14-amino acid sequence of Cap43 protein, TRSRSHTSEGTRSR. J. Inorg. Biochem. 2001, 84, 47–54. [Google Scholar] [CrossRef]
- Karavelas, T.; Malandrinos, G.; Hadjiliadis, N.; Mlynarz, P.; Kozlowski, H.; Barsan, M.; Butler, I. Coordination properties of Cu(II) and Ni(II) ions towards the C-terminal peptide fragment -TYTEHA- of histone H4. Dalton. Trans. 2008, 9, 1215–1223. [Google Scholar]
- Gajda, T.; Henry, B.; Aubry, A.; Delpuech, J.-J. Proton and metal-ion interactions with glycyl-glycyl-histamine mimicking Serum Albumin. Inorg. Chem. 1996, 35, 586–593. [Google Scholar] [CrossRef]
- Mylonas, M.; Plakatouras, J.C.; Hadjiliadis, N.; Papavasileiou, K.D.; Melissas, V.S. An extremely stable Ni(II) complex derived from the hydrolytic cleavage of the C-terminal tail of histone H2A. J. Inorg. Biochem. 2005, 99, 637–643. [Google Scholar] [CrossRef]
- Bal, W.; Jezowska-Bojczuk, M.; Kasprzak, K.S. Binding of nickel(II) and copper(II) to the N-terminal sequence of human protamine HP2. Chem. Res. Toxicol. 1997, 10, 906–914. [Google Scholar] [CrossRef]
- Hay, R.W.; Hassan, M.M.; You-Quan, C. Kinetic and thermodynamic studies of the copper (II) and nickel(II) complexes of glycylglycyl-L-histidine. J. Inorg. Biochem. 1993, 52, 17–25. [Google Scholar]
- Raycheba, J.M.T.; Margerum, D.W. Effect of noncoordinative axial blocking on the stability and kinetic behavior of ternary 2,6-lutidine-nickel(II)-oligopeptide complexes. Inorg. Chem. 1980, 19, 837–843. [Google Scholar] [CrossRef]
- Yamashita, M.M.; Wesson, L.; Eisenman, G.; Eisenberg, D. Where metal ions bind in proteins. Proc. Natl. Acad. Sci. USA 1990, 87, 5648–5652. [Google Scholar] [CrossRef]
- Regan, L. The design of metal-binding sites in proteins. Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 257–287. [Google Scholar] [CrossRef]
- Zoroddu, M.A.; Peana, M.; Medici, S.; Casella, L.; Monzani, E.; Costa, M. Nickel binding to histone H4. Dalton. Trans. 2010, 39, 787–793. [Google Scholar] [CrossRef]
- Peana, M.; Medici, S.; Nurchi, V.M.; Crisponi, G.; Zoroddu, M.A. Nickel binding sites in histone proteins: Spectroscopic and structural characterization. Coord. Chem. Rev. 2013, 257, 2737–2751. [Google Scholar] [CrossRef]
- Zoroddu, M.A.; Peana, M.; Medici, S. Multidimensional NMR spectroscopy for the study of histone H4-Ni(II) interaction. Dalton. Trans. 2007, 3, 379–384. [Google Scholar] [CrossRef]
- Bal, W.; Lukszo, J.; Bialkowski, K.; Kasprzak, K.S. Interactions of Nickel(II) with histones: interactions of Nickel(II) with CH3CO-Thr-Glu-Ser-His-His-Lys-NH2, a peptide modeling the potential metal binding site in the “C-Tail” region of histone H2A. Chem. Res. Toxicol. 1998, 11, 1014–1023. [Google Scholar] [CrossRef]
- Bal, W.; Liang, R.; Lukszo, J.; Lee, S.H.; Dizdaroglu, M.; Kasprzak, K.S. Ni(II) specifically cleaves the C-terminal tail of the major variant of histone H2A and forms an oxidative damage-mediating complex with the cleaved-off octapeptide. Chem. Res. Toxicol. 2000, 13, 616–624. [Google Scholar] [CrossRef]
- Mlynarz, P.; Gaggelli, N.; Panek, J.; Stasiak, M.; Valensin, G.; Kowalik-Jankowska, T.; Leplawy, M.T.; Latajka, Z.; Kozlowski, H. How the α-hydroxymethylserine residue stabilizes oligopeptide complexes with nickel(II) and copper(II) ions. J. Chem. Soc. Dalton. Trans. 2000, 1033–1038. [Google Scholar]
- Nunes, A.M.; Zavitsanos, K.; Del Conte, R.; Malandrinos, G.; Hadjiliadis, N. Interaction of histone H2B (fragment 63-93) with Ni(ii). An NMR study. Dalton. Trans. 2009, 11, 1904–1913. [Google Scholar]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Valensin, D.; Gajda, K.; Gralka, E.; Valensin, G.; Kamysz, W.; Kozlowski, H. Copper binding to chicken and human prion protein amylodogenic regions: differences and similarities revealed by Ni2+ as a diamagnetic probe. J. Inorg. Biochem. 2010, 104, 71–78. [Google Scholar] [CrossRef]
- Kozlowski, H.; Bal, W.; Dyba, M.; Kowalik-Jankowska, T. Specific structure-stability relations in metallopeptides. Coord. Chem. Rev. 1999, 184, 319–346. [Google Scholar] [CrossRef]
- Bal, W.; Lukszo, J.; Kasprzak, K.S. Mediation of oxidative DNA damage by nickel(II) and copper(II) complexes with the N-terminal sequence of human protamine HP2. Chem. Res. Toxicol. 1997, 10, 915–921. [Google Scholar] [CrossRef]
- Liang, R.; Senturker, S.; Shi, X.; Bal, W.; Dizdaroglu, M.; Kasprzak, K.S. Effects of Ni(II) and Cu(II) on DNA interaction with the N-terminal sequence of human protamine P2: enhancement of binding and mediation of oxidative DNA strand scission and base damage. Carcinogenesis 1999, 20, 893–898. [Google Scholar] [CrossRef]
- Bunin, G.R.; Noller, K.; Rose, P.; Smith, E. Carcinogenesis. In Occupational and Reproductive Hazards: A Guide for Clinicians; Paul, M., Ed.; Williams and Wilkins: Baltimore, MD, USA, 1992; pp. 76–88. [Google Scholar]
- Brunger, A.T. X-PLOR, version 3.1; Howard Hughes Medical Institute and Yale University: New Haven, CT, USA, 1993. [Google Scholar]
- Bal, W.; Djuran, M.I.; Margerum, D.W.; Gray, E.T., Jr.; Mazid, M.A.; Tom, R.T.; Nieboer, E.; Sadler, P.J. Dioxygen-induced decarboxylation and hydroxylation of [Ni(II)(Glycyl-Glycyl-LHistidine)] occurs via Ni(III): X-ray crystal structure of [Ni(II)(Glycyl-Glycyl-α- hydroxy-D,L- Histamine).3H2O)]. J. Chem. Soc. Chem. Commun. 1994, 1889–1890. [Google Scholar]
- Zhou, D.; Salnikow, K.; Costa, M. Cap43, a novel gene specifically induced by Ni2+ compounds. Cancer Res. 1998, 58, 2182–2189. [Google Scholar]
- Salnikow, K.; Kluz, T.; Costa, M. Role of Ca2+ in the regulation of nickel-inducible Cap43 gene expression. Toxicol. Appl. Pharmacol. 1999, 160, 127–132. [Google Scholar] [CrossRef]
- Nishie, A.; Masuda, K.; Otsubo, M.; Migita, T.; Tsuneyoshi, M.; Kohno, K.; Shuin, T.; Naito, S.; Ono, M.; Kuwano, M. High expression of the Cap43 gene in infiltrating macrophages of human renal cell carcinomas. Clin. Cancer Res. 2001, 7, 2145–2151. [Google Scholar]
- Nishio, S.; Ushijima, K.; Tsuda, N.; Takemoto, S.; Kawano, K.; Yamaguchi, T.; Nishida, N.; Kakuma, T.; Tsuda, H.; Kasamatsu, T.; et al. Cap43/NDRG1/Drg-1 is a molecular target for angiogenesis and a prognostic indicator in cervical adenocarcinoma. Cancer Lett. 2008, 264, 36–43. [Google Scholar] [CrossRef]
- Zoroddu, M.A.; Peana, M.; Medici, S.; Anedda, R. An NMR study on nickel binding sites in Cap43 protein fragments. Dalton. Trans. 2009, 28, 5523–5534. [Google Scholar]
- Denkhaus, E.; Salnikow, K. Nickel essentiality, toxicity, and carcinogenicity. Crit. Rev. Oncol. Hematol. 2002, 42, 35–56. [Google Scholar] [CrossRef]
- Alagna, L.; Hasnain, S.S.; Piggott, B.; Williams, D.J. The nickel ion environment in jack bean urease. Biochem. J. 1984, 220, 591–595. [Google Scholar]
- Vollmer, J.; Fritz, M.; Dormoy, A.; Weltzien, H.U.; Moulon, C. Dominance of the BV17 element in nickel-specific human T cell receptors relates to severity of contact sensitivity. Eur. J. Immunol. 1997, 27, 1865–1874. [Google Scholar] [CrossRef]
- Rothenberg, M.E. Innate sensing of nickel. Nat. Immunol. 2010, 11, 781–782. [Google Scholar] [CrossRef]
- Jones, C.E.; Klewpatinond, M.; Abdelraheim, S.R.; Brown, D.R.; Viles, J.H. Probing copper2+ binding to the prion protein using diamagnetic nickel2+ and 1H NMR: the unstructured N terminus facilitates the coordination of six copper2+ ions at physiological concentrations. J. Mol. Biol. 2005, 346, 1393–1407. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Delogu, L.G.; Zoroddu, M.A. Mn(II) and Zn(II) interactions with peptide fragments from Parkinson's disease genes. Dalton. Trans. 2012, 41, 4378–4388. [Google Scholar] [CrossRef]
- Remelli, M.; Peana, M.; Medici, S.; Delogu, L.G.; Zoroddu, M.A. Interaction of divalent cations with peptide fragments from Parkinson's disease genes. Dalton. Trans. 2013, 42, 5964–5974. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Medici, S.; Peana, M.; Nurchi, V.M.; Zoroddu, M.A. The Involvement of Amino Acid Side Chains in Shielding the Nickel Coordination Site: An NMR Study. Molecules 2013, 18, 12396-12414. https://doi.org/10.3390/molecules181012396
Medici S, Peana M, Nurchi VM, Zoroddu MA. The Involvement of Amino Acid Side Chains in Shielding the Nickel Coordination Site: An NMR Study. Molecules. 2013; 18(10):12396-12414. https://doi.org/10.3390/molecules181012396
Chicago/Turabian StyleMedici, Serenella, Massimiliano Peana, Valeria Marina Nurchi, and Maria Antonietta Zoroddu. 2013. "The Involvement of Amino Acid Side Chains in Shielding the Nickel Coordination Site: An NMR Study" Molecules 18, no. 10: 12396-12414. https://doi.org/10.3390/molecules181012396
APA StyleMedici, S., Peana, M., Nurchi, V. M., & Zoroddu, M. A. (2013). The Involvement of Amino Acid Side Chains in Shielding the Nickel Coordination Site: An NMR Study. Molecules, 18(10), 12396-12414. https://doi.org/10.3390/molecules181012396