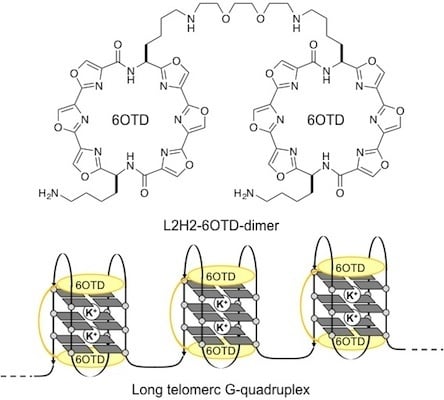
Evaluation of the Interaction between Long Telomeric DNA and Macrocyclic Hexaoxazole (6OTD) Dimer of a G-quadruplex Ligand
Abstract
:1. Introduction



2. Results and Discussion



| DNA oligomers | Ligands | Tm |
|---|---|---|
| absence | 59 | |
| telo24 | 2 (4 eq.) | 75 |
| 3 (2 eq.) | 73 | |
| absence | 57 | |
| telo48 | 2 (8 eq.) | 76 |
| 3 (4 eq.) | 74 | |
| absence | 55 | |
| telo48 | 2 (12 eq.) | 73 |
| 3 (6 eq.) | 70 | |
| absence | 55 | |
| telo48 | 2 (16 eq.) | 70 |
| 3 (8 eq.) | 69 |
3. Experimental
3.1. General
3.2. Synthesis
3.3. Electrophoresis Mobility Shift Assay (EMSA)
3.4. CD Spectrometry
3.5. CD Melting Experiments
3.6. Telomerase Repeat Amplification Protocol (TRAP) Assay
4. Conclusions
Supplementary Materials
Acknowledgments
References and Notes
- Blackburn, E.H. Switching and Signaling at the Telomere. Cell 2001, 106, 661–673. [Google Scholar] [CrossRef]
- Hemann, M.T.; Greider, C.W. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 1999, 27, 3964–3969. [Google Scholar] [CrossRef]
- de Lange, T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef]
- de Lange, T. How Shelterin solves the telomere end-protection problem. Cold Spring Harbor Symp. Quant. Biol. 2010, 75, 167–177. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Qi, H.; Liu, L.F. Protection of DNA Ends by Telomeric 3’ G-Tail Sequences. J. Biol. Chem. 2007, 282, 18786–18792. [Google Scholar] [CrossRef]
- Davis, J.T. G-Quartets 40 years later: From 5'-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004, 43, 668–698. [Google Scholar] [CrossRef]
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef]
- Neidle, S.; Parkinson, G.N. Quadruplex DNA crystal structures and drug. Biochimie 2008, 90, 1184–1196. [Google Scholar] [CrossRef]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010, 277, 1107–1117. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar]
- Shay, J.W.; Wright, W.E. Telomerase therapeutics for cancer: challenges and new directions. Nat. Rev. Drug Discov. 2006, 5, 577–584. [Google Scholar] [CrossRef]
- Zahler, A.M.; Williamson, J.R.; Cech, T.R.; Prescott, D.M. Inhibition of telomerase by G-quartet DNA structures. Nature 1991, 350, 718–720. [Google Scholar] [CrossRef]
- Fumagalli, M.; Rossiello, F.; Clerici, M.; Barozzi, S.; Cittaro, D.; Kaplunov, J.M.; Bucci, G.; Dobreva, M.; Matti, V.; Beausejour, C.M.; et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 2012, 14, 335–365. [Google Scholar] [CrossRef]
- Neidle, S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010, 277, 1118–1125. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.-P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar]
- Ou, T.M.; Lu, Y.J.; Tan, J.H.; Huang, Z.S.; Wong, K.Y.; Gu, L.Q. G-Quadruplexes: Targets in Anticancer Drug Design. ChemMedChem 2008, 3, 690–713. [Google Scholar]
- Shin-ya, K.; Wierzba, K.; Matsuo, K.; Ohtani, T.; Yamada, Y.; Furihata, K.; Hayakawa, Y.; Seto, H. Telomestatin, a Novel Telomerase Inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 2001, 123, 1262–1263. [Google Scholar] [CrossRef]
- Kim, M.Y.; Vankayalapati, H.; Shin-Ya, K.; Wierzba, K.; Hurley, L.H. Telomestatin, a Potent Telomerase Inhibitor That Interacts Quite Specifically with the Human Telomeric Intramolecular G-Quadruplex. J. Am. Chem. Soc. 2002, 124, 2098–2099. [Google Scholar]
- Kim, M.Y.; Gleason-Guzman, M.; Izbicka, E.; Nishioka, D.; Hurley, L.H. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-Quadruplex structures. Cancer Res. 2003, 63, 3247–3256. [Google Scholar]
- Rosu, F.; Gabelica, V.; Shin-ya, K.; Pauw, E.D. Telomestatin-induced stabilization of the human telomeric DNA quadruplex monitored by electrospray mass spectrometry. Chem. Commun. 2003, 2702–2703. [Google Scholar]
- Rezler, E.M.; Seenisamy, J.; Bashyam, S.; Kim, M.Y.; White, E.; Wilson, W.D.; Hurley, L.H. Telomestatin and Diseleno Sapphyrin Bind Selectively to Two Different Forms of the Human Telomeric G-Quadruplex Structure. J. Am. Chem. Soc. 2005, 127, 9439–9447. [Google Scholar]
- Doi, T.; Shibata, K.; Yoshida, M.; Takagi, M.; Tera, M.; Nagasawa, K.; Shin-ya, K.; Takahashi, T. (S)-Stereoisomer of telomestatin as a potent G-quadruplex binder and telomerase inhibitor. Org. Biomol. Chem. 2011, 9, 387–393. [Google Scholar] [CrossRef]
- Minhas, G.S.; Plich, D.S.; Kerrigan, J.E.; LaVoie, E.J.; Rice, J.E. Synthesis and G-quadruplex stabilizing properties of a series of oxazole-containing macrocycles. Bioorg. Med. Chem. Lett. 2006, 16, 3891–3895. [Google Scholar] [CrossRef]
- Barbieri, C.M.; Srinivasan, A.R.; Rzuczek, S.G.; Rice, J.E.; LaVoie, E.J.; Pilch, D.S. Defining the mode, energetics and specificity with which a macrocyclic hexaoxazole binds to human telomeric G-quadruplex DNA. Nucleic Acids Res. 2007, 35, 3272–3286. [Google Scholar]
- Rzuczek, S.G.; Pilch, D.S.; LaVoie, E.J.; Rice, J.E. Lysinyl macrocyclic hexaoxazoles: Synthesis and selective G-quadruplex stabilizing properties. Bioorg. Med. Chem. Lett. 2008, 18, 913–917. [Google Scholar] [CrossRef]
- Satyanarayana, M.; Rzuczek, S.G.; LaVoie, E.J.; Pilch, D.S.; Liu, A.; Liu, L.F.; Rice, J.E. Ring-closing metathesis for the synthesis of a highly G-quadruplex selective macrocyclic hexaoxazole having enhanced cytotoxic potency. Bioorg. Med. Chem. Lett. 2008, 18, 3802–3804. [Google Scholar]
- Pilch, D.S.; Barbieri, C.M.; Rzuczek, S.G.; LaVoie, E.J.; Rice, J.E. Targeting human telomeric G-quadruplex DNA with oxazole-containing macrocyclic compounds. Biochimie 2008, 90, 1233–1249. [Google Scholar]
- Tsai, Y.C.; Qi, H.; Lin, C.P.; Lin, R.K.; Kerrigan, J.E.; Rzuczek, S.G.; LaVoie, E.J.; Rice, J.E.; Pilch, D.S.; Lyu, Y.L.; et al. A G-quadruplex Stabilizer Induces M-phase Cell Cycle Arrest. J. Biol. Chem. 2009, 284, 22535–22543. [Google Scholar] [CrossRef]
- Rzuczek, S.G.; Pilch, D.S.; Liu, A.; Liu, L.; LaVoie, E.J.; Rice, J.E. Macrocyclic Pyridyl Polyoxazoles: Selective RNA and DNA G-Quadruplex Ligands as Antitumor Agents. J. Med. Chem. 2010, 53, 3632–3644. [Google Scholar] [CrossRef]
- Satyanarayana, M.; Kim, Y.A.; Rzuczek, S.G.; Pilch, D.S.; Liu, A.A.; Liu, L.F.; Rice, J.E.; LaVoie, E.J. Macrocyclic hexaoxazoles: Influence of aminoalkyl substituents on RNA and DNA G-quadruplex stabilization and cytotoxicity. Bioorg. Med. Chem. Lett. 2010, 20, 3150–3154. [Google Scholar] [CrossRef]
- Tera, M.; Sohtome, Y.; Ishizuka, H.; Doi, T.; Takagi, M.; Shin-ya, K.; Nagasawa, K. Design and synthesis of telomestatin derivatives and their inhibitory activity of telomerase. Heterocycles 2006, 69, 505–514. [Google Scholar] [CrossRef]
- Tera, M.; Ishizuka, H.; Takagi, M.; Suganuma, M.; Shin-ya, K.; Nagasawa, K. Macrocyclic Hexaoxazoles as Sequence- and Mode-Selective G-Quadruplex Binders. Angew. Chem. Int. Ed. 2008, 47, 5557–5560. [Google Scholar] [CrossRef]
- Majima, S.; Tera, M.; Iida, K.; Shin-ya, K.; Nagasawa, K. Design and synthesis of telomestatin derivatives containing Methyl Oxazole and their G-Quadruplex stabilizing activities. Heterocycles 2011, 82, 1345–1357. [Google Scholar]
- Iida, K.; Majima, S.; Ohtake, T.; Tera, M.; Shin-ya, K.; Nagasawa, K. Design and synthesis of G-Quadruplex ligands bearing macrocyclic hexaoxazoles with four-way side chains. Heterocycles 2012, 84, 401–411. [Google Scholar] [CrossRef]
- Nakamura, T.; Iida, K.; Tera, M.; Shin-ya, K.; Seimiya, H.; Nagasawa, K. A Caged Ligand for a Telomeric G-Quadruplex. ChemBioChem 2012, 13, 774–777. [Google Scholar] [CrossRef]
- Iida, K.; Tera, M.; Hirokawa, T.; Shin-ya, K.; Nagasawa, K. G-quadruplex recognition by macrocyclic hexaoxazole (6OTD) dimer: greater selectivity than monomer. Chem. Commun. 2009, 6481–6483. [Google Scholar]
- Iida, K.; Tera, M.; Hirokawa, T.; Shin-ya, K.; Nagasawa, K. Synthesis of Macrocyclic Hexaoxazole (6OTD) Dimers, Containing Guanidine and Amine Functionalized Side Chains, and an Evaluation of Their Telomeric G4 Stabilizing Properties. J. Nucleic Acids 2010, 2010. 217627. [Google Scholar]
- Amemiya, Y.; Furunaga, Y.; Iida, K.; Tera, M.; Nagasawa, K.; Ikebukuro, K.; Nakamura, C. Analysis of the unbinding force between telomestatin derivatives and human telomeric G-quadruplex by atomic force microscopy. Chem. Commun. 2011, 47, 7485–7487. [Google Scholar] [CrossRef]
- Tera, M.; Iida, K.; Ishizuka, H.; Takagi, M.; Suganuma, M.; Doi, T.; Shin-ya, K.; Nagasawa, K. Synthesis of a Potent G-Quadruplex-Binding Macrocyclic Heptaoxazole. ChemBioChem 2009, 10, 431–435. [Google Scholar]
- Tera, M.; Iida, K.; Ikebukuro, K.; Seimiya, H.; Shin-ya, K.; Nagasawa, K. Visualization of G-quadruplexes by using a BODIPY-labeled macrocyclic heptaoxazole. Org. Biomol. Chem. 2010, 8, 2749–2755. [Google Scholar] [CrossRef]
- We named the core structure of macrocyclic hexaoxazole of 2 as 6OTD (6 Oxazoles Telomestatin Derivative).
- Xu, Y.; Ishizuka, T.; Kurabayashi, K.; Komiyama, M. Consecutive formation of G-quadruplexes in human telomeric-overhang DNA: A protective capping structure for telomere ends. Angew. Chem. Int. Ed. 2009, 48, 7833–7836. [Google Scholar] [CrossRef]
- Petraccone, L.; Trent, J.O.; Chaires, J.B. The tail of the telomere. J. Am. Chem. Soc. 2008, 130, 16530–16532. [Google Scholar] [CrossRef]
- Renciuk, D.; Kejnovska, I.; Skolakova, P.; Bednarova, K.; Motlova, J.; Vorlıckova, M. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009, 37, 6625–6634. [Google Scholar]
- Vorlıckova, M.; Chladkova, J.; Kejnovska, I.; Fialova, M.; Kypr, J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005, 33, 5851–5860. [Google Scholar]
- Singh, V.; Azarkh, M.; Drescher, M.; Hartig, J. S. Conformations of individual quadruplex units studied in the context of extended human telomeric DNA. Chem. Commun. 2012, 48, 8258–8260. [Google Scholar] [CrossRef]
- Petraccone, L.; Spink, C.; Trent, J.O.; Garbett, N.C.; Mekmaysy, C.S.; Giancola, C.; Chaires, J.B. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011, 133, 20951–20961. [Google Scholar]
- Yu, H.Q.; Miyoshi, D.; Sugimoto, N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006, 128, 15461–15468. [Google Scholar] [CrossRef]
- Yu, H.; Gu, X.; Nakano, S.; Miyoshi, D.; Sugimoto, N. Beads-on-a-string structure of long telomeric DNAs under molecular crowding conditions. J. Am. Chem. Soc. 2012, 134, 20060–20069. [Google Scholar] [CrossRef]
- Haider, S.; Parkinson, G.N.; Neidle, S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008, 95, 296–311. [Google Scholar] [CrossRef]
- Haider, S.; Neidle, S. A molecular model for drug binding to tandem repeats of telomeric G-quadruplexes. Biochem. Soc. Trans. 2009, 37, 583–588. [Google Scholar] [CrossRef]
- Chang, C.C.; Chien, C.W.; Lin, Y.H.; Kang, C.C.; Chang, T.C. Investigation of spectral conversion of d(TTAGGG)4 and d(TTAGGG)13 upon potassium titration by a G-quadruplex recognizer BMVC molecule. Nucleic Acids Res. 2007, 35, 2846–2860. [Google Scholar]
- Iida, K.; Tsubouchi, G.; Nakamura, T.; Majima, S.; Seimiya, H.; Nagasawa, K. Interaction of long telomeric DNAs with macrocyclic hexaoxazole as a G-quadruplex ligand. Med. Chem. Commun. 2013, 4, 260–264. [Google Scholar] [CrossRef]
- The dimer 3 was found to form a complex with telo24 in 1:1 ratio based upon the electrospray ionization mass spectra (see Supplementary Information).
- We prepared long telomeric DNAs consisting of 48, 72, and 96 bases, i.e., (TTAGGG)n (n = 8, 12, 16; telo48, telo72, and telo96, corresponding to two, three and four units of telo24, respectively). The DNAs were purchased from Sigma Genosys. See table below for sequences of telomeric DNAs used in this work.
- Balagurumoorthy, P.; Brahmachari, S.K. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994, 269, 21858–21869. [Google Scholar]
- TRAP lysis buffer was prepared as follows; 400 μL of 1 M Tris–HCl (pH 8.0), 40 μL of 1 M MgCl2 aq, 200 μL of 0.2 M EGTA, 2 mL of 10% CHAPS, 8 mL of 50% glycerol and 25 mL of RNase free H2O were mixed. Next, HCl aq was added to the mixture (final; pH 7.5), which was diluted to 40 mL with MilliQ water and filtered. Then, 1.0 μL of 0.1 M 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) and 0.35 μL of 2-mercaptoethanol were added to 1.0 mL of the mixture.
- Sample Availability: Samples of the compounds L2H2-6OTD (2) and L2H2-6OTD-dimer (3) are available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Iida, K.; Majima, S.; Nakamura, T.; Seimiya, H.; Nagasawa, K. Evaluation of the Interaction between Long Telomeric DNA and Macrocyclic Hexaoxazole (6OTD) Dimer of a G-quadruplex Ligand. Molecules 2013, 18, 4328-4341. https://doi.org/10.3390/molecules18044328
Iida K, Majima S, Nakamura T, Seimiya H, Nagasawa K. Evaluation of the Interaction between Long Telomeric DNA and Macrocyclic Hexaoxazole (6OTD) Dimer of a G-quadruplex Ligand. Molecules. 2013; 18(4):4328-4341. https://doi.org/10.3390/molecules18044328
Chicago/Turabian StyleIida, Keisuke, Satoki Majima, Takahiro Nakamura, Hiroyuki Seimiya, and Kazuo Nagasawa. 2013. "Evaluation of the Interaction between Long Telomeric DNA and Macrocyclic Hexaoxazole (6OTD) Dimer of a G-quadruplex Ligand" Molecules 18, no. 4: 4328-4341. https://doi.org/10.3390/molecules18044328
APA StyleIida, K., Majima, S., Nakamura, T., Seimiya, H., & Nagasawa, K. (2013). Evaluation of the Interaction between Long Telomeric DNA and Macrocyclic Hexaoxazole (6OTD) Dimer of a G-quadruplex Ligand. Molecules, 18(4), 4328-4341. https://doi.org/10.3390/molecules18044328