Structural Characterization by NMR of a Double Phosphorylated Chimeric Peptide Vaccine for Treatment of Alzheimer’s Disease
Abstract
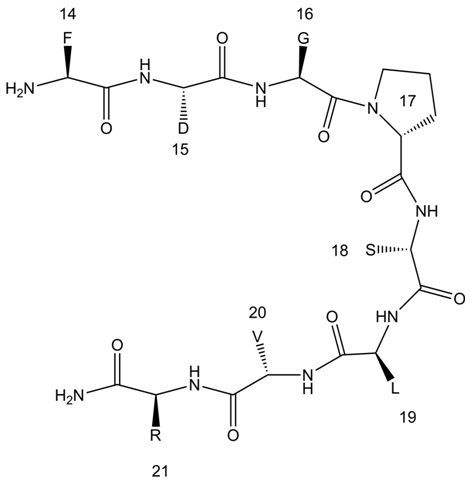
:1. Introduction
2. Results and Discussion





3. Experimental
3.1. Peptide Synthesis
3.2. NMR Spectroscopy
3.3. Statistical Analysis

3.4. 3JHN-Hα Couplings Analysis

4. Conclusions
Supplementary Materials
Acknowledgments
References
- Mei, C.G.; Jahr, N.; Singer, D.; Berger, S. Hairpin conformation of an 11-mer peptide. Bioorg. Med. Chem. 2011, 19, 3497–3501. [Google Scholar]
- Paine, G.H.; Scheraga, H.A. Prediction of the native conformation of a polypeptide by a statistical-mechanical procedure. I. Backbone structure of enkephalin. Biopolymers 1985, 24, 1391–1436. [Google Scholar] [CrossRef]
- Blanco, F.J.; Jimenez, M.A.; Herranz, J.; Rico, M.; Santoro, J.; Nieto, J.L. NMR evidence of a short linear peptide that folds into a .beta.-hairpin in aqueous solution. J. Am. Chem. Soc. 1993, 115, 5887–5888. [Google Scholar]
- Wright, P.E.; Dyson, H.J.; Lerner, R.A. Conformation of peptide fragments of proteins in aqueous solution: Implications for initiation of protein folding. Biochemistry 1988, 27, 7167–7175. [Google Scholar] [CrossRef]
- Mittag, T.; Forman-Kay, J.D. Atomic-level characterization of disordered protein ensembles. Curr. Opin. Struct. Biol. 2007, 17, 3–14. [Google Scholar] [CrossRef]
- Eliezer, D. Biophysical characterization of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2009, 19, 23–30. [Google Scholar] [CrossRef]
- Fesinmeyer, R.M.; Hudson, F.M.; Olsen, K.A.; White, G.W.N.; Euser, A.; Andersen, N.H. Chemical shifts provide fold populations nd register of beta hairpins and beta sheets. J. Biomol. NMR 2005, 33, 213–231. [Google Scholar] [CrossRef]
- Gross, K.-H.; Kalbitzer, H.R. Distribution of chemical shifts in 1H nuclear magnetic resonance spectra of proteins. J. Magn. Reson. 1988, 76, 87–99. [Google Scholar]
- Spera, S.; Bax, A. Empirical correlation between protein backbone conformation and C.alpha. and C.beta. 13C nuclear magnetic resonance chemical shifts. J. Am. Chem. Soc. 1991, 113, 5490–5492. [Google Scholar] [CrossRef]
- Szilagyi, L.; Jardetzky, O. α-Proton chemical shifts and secondary structure in proteins. J. Magn. Reson. 1989, 83, 441–449. [Google Scholar]
- Meier, S.; Blackledge, M.; Grzesiek, S. Conformational distributions of unfolded polypeptides from novel NMR techniques. J. Chem. Phys. 2008, 128, 052204. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Linking Folding and Binding. Curr. Opin. Struct. Biol. 2009, 19, 31–38. [Google Scholar] [CrossRef]
- Jensen, M.R.; Markwick, P.; Griesinger, C.; Zweckstetter, M.; Meier, S.; Grzesiek, S.; Bernado, P.; Blackledge, M. Struct. Bonding 2009, 17, 1169–1185. [CrossRef]
- Hutchinson, E.G.; Thornton, J.M. A revised set of potentials for beta turn formation in proteins. Protein Sci. 1994, 3, 2207–2216. [Google Scholar] [CrossRef]
- Rose, G.D.; Glerasch, L.M.; Smith, J.A. Turns in Peptides and Proteins. Adv. Protein Chem. 1985, 37, 1–109. [Google Scholar] [CrossRef]
- Wilmot, C.M.; Thornton, J.M. Analysis and prediction of the different types of β-turn in proteins. J. Mol. Biol. 1988, 203, 221–232. [Google Scholar] [CrossRef]
- Kaumaya, P.T.; Foy, K.C.; Garrett, J.; Rawale, S.V.; Vicari, D.; Thurmond, J.M.; Lamb, T.; Mani, A.; Kane, Y.; Balint, C.R.; et al. Phase I active immunotherapy with combination of two chimeric, human epidermal growth factor receptor 2, B-cell epitopes fused to a promiscuous T-cell epitope in patients with metastatic and/or recurrent solid tumors. J. Clin. Oncol. 2009, 27, 5270–5277. [Google Scholar] [CrossRef]
- Menéndez-González, M.; Pérez-Piñera, P.; Martínez-Rivera, M.; Muñiz, A.L.; Vega, J.A. Immunotherapy for Alzheimer’s disease: Rational basis in ongoing clinical trials. Curr. Pharm. Des. 2011, 17, 508–520. [Google Scholar] [CrossRef]
- Wolf, A.; Mozdzanowska, K.; Williams, K.L.; Singer, D.; Richter, M.; Hoffmann, R.; Caton, A.J.; Otvos, L.; Erikson, J. Vaccination with M2e-based multiple antigenic peptides: Characterization of the B cell response and protection efficacy in inbred and outbred mice. PLoS One 2011, 6, e28445. [Google Scholar] [CrossRef]
- Citron, M. Alzheimer’s disease: Strategies for disease modification. Nat. Rev. Drug Discov. 2010, 9, 387–398. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging 1995, 16, 271–278. [Google Scholar] [CrossRef]
- Shah, S.; Federoff, H.J. Therapeutic potential of vaccines for Alzheimer’s disease. Immunotherapy 2011, 3, 287–298. [Google Scholar] [CrossRef]
- Weiner, H.L.; Frenkel, D. Immunology and immunotherapy of Alzheimer’s disease. Nat. Rev. Immunol. 2006, 6, 404–416. [Google Scholar]
- Asuni, A.A.; Boutajangout, A.; Quartermain, D.; Sigurdsson, E.M. Immunotherapy Targeting Pathological Tau Conformers in a Tangle Mouse Model Reduces Brain Pathology with Associated Functional Improvements. J. Neurosci. 2007, 27, 9115–9129. [Google Scholar] [CrossRef]
- Sigurdsson, E.M.; Knudsen, E.; Asuni, A.; Fitzer-Attas, C.; Sage, D.; Quartermain, D.; Goni, F.; Frangione, B.; Wisniewski, T. An attenuated immune response is sufficient to enhance cognition in an Alzheimer’s disease mouse model immunized with Amyloid-β derivatives. J. Neurosci. 2004, 24, 6277–6282. [Google Scholar] [CrossRef]
- Dakappagari, N.K.; Douglas, D.B.; Triozzi, P.L.; Stevens, V.C.; Kaumaya, P.T.P. Prevention of Mammary Tumors with a Chimeric HER-2 B-cell Epitope Peptide Vaccine. Cancer Res. 2000, 60, 3782–3789. [Google Scholar]
- Kaumaya, P.T.; Berndt, K.D.; Heidorn, D.B.; Trewhella, J.; Kezdy, F.J.; Goldberg, E. Synthesis and biophysical characterization of engineered topographic immunogenic determinants with alpha alpha topology. Biochemistry (Wash.) 1990, 29, 13–23. [Google Scholar] [CrossRef]
- Lairmore, M.D.; DiGeorge, A.M.; Conrad, S.F.; Trevino, A.V.; Lal, R.B.; Kaumaya, P.T. Human T-lymphotropic virus type 1 peptides in chimeric and multivalent constructs with promiscuous T-cell epitopes enhance immunogenicity and overcome genetic restriction. J. Virol. 1995, 69, 6077–6089. [Google Scholar]
- Singer, D.; Lehmann, J.; Hanisch, K.; Hartig, W.; Hoffmann, R. Neighbored phosphorylation sites as PHF-tau specific markers in Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2006, 346, 819–828. [Google Scholar] [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Available online: http://deposit.bmrb.wisc.edu/author_view/BMRB/19112_hy_errejbgi.str (accessed on 19 April 2013).
- Case, D.A.; Dyson, H.; Wright, P.E. Use of chemical shifts and coupling constants in nuclear magnetic resonance structural studies on peptides and proteins. Methods Enzymol. 1994, 239, 392–416. [Google Scholar] [CrossRef]
- Sharman, G.J.; Griffiths-Jones, S.R.; Jourdan, M.; Searle, M.S. Effects of the amino acid (phi, psi) propensities and secondary structure interactions in modulatin Halfa shift in peptide ans protein beta-sheet. J. Am. Chem. Soc 2001, 123, 12318–12324. [Google Scholar] [CrossRef]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar] [CrossRef]
- Santiveri, C.; Rico, M.; Jiménez, M. 13Cα and 13Cβ chemical shifts as a tool to delineate β-hairpin structures in peptides. J. Biomol. NMR 2001, 19, 331–345. [Google Scholar] [CrossRef]
- Wishart, D.S.; Sykes, B.D. Chemical-Shifts As A Tool for Structure Determination. Nuclear Magnetic Resonance Pt. C 1994, 239, 363–392. [Google Scholar] [CrossRef]
- Bundi, A.; Wuthrich, K. H-1-Nmr parameters of the common amino-acid residues measured in aqueous-solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-OH. Biopolymers (Biospectrosc.) 1979, 18, 285–297. [Google Scholar] [CrossRef]
- Modig, K.; Jurgensen, V.W.; Lindorff-Larsen, K.; Fieber, W.; Bohr, H.G.; Poulsen, F.M. Detection of initiation sites in protein folding of the four helix bundle ACBP by chemical shift analysis. FEBS Lett. 2007, 581, 4965–4971. [Google Scholar]
- Karplus, M. Vicinal proton coupling in nuclear magnetic resonance. J. Am. Chem. Soc. 1963, 85, 2870–2871. [Google Scholar] [CrossRef]
- Gomori, G. Preparation of buffers for use in enzyme studies. Methods Enzymol. 1955, 1, 138–146. [Google Scholar] [CrossRef]
- Sørensen, S.P.L. Biochem. Z. 1909, 21, 131.
- Biological Magnetic Resonance Data Bank. Available online: http://www.bmrb.wisc.edu/ref_info/statsel.htm/ (accessed on 19 April 2013).
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, D402–D408. [Google Scholar]
- Schwarzinger, S.; Kroon, G.J.A.; Foss, T.R.; Wright, P.E.; Dyson, H.J. Random coil chemical shifts in acidic 8 M urea: Implementation of random coil shift data in NMRView. J. Biomol NMR 2000, 18, 43–48. [Google Scholar] [CrossRef]
- Cavanagh, J.; Fairbrother, W.J.; Palmer, A.G., III; Rance, M.; Skelton, N.J. Protein NMR Spectroscopy; Academic Press Inc.: San Diego, CA, USA, 2006. [Google Scholar]
- Ozenne, V.; Bauer, F.; Salmon, L.; Huang, J.R.; Jensen, M.R.; Segard, S.; Bernado, P.; Charavay, C.; Backledge, M. Flexible-meccano: A tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics 2012, 28, 1463–1470. [Google Scholar] [CrossRef]
- Smith, L.J.; Bolin, K.A.; Schwalbe, H.; MacArthur, W.M.; Thornton, J.M.; Dobson, C.M. Analysis of main chain torsion angles in proteins: Prediction of NMR coupling constants for native and random coil conformations. J. Mol. Biol. 1996, 255, 494–506. [Google Scholar] [CrossRef]
- Bielska, A.A.; Zondlo, N.J. Hyperphosphorylation of tau induces local polyproline II helix. Biochemistry (Wash.) 2006, 45, 5527–5537. [Google Scholar] [CrossRef]
- D'Souza, S.; Rosseels, V.R.A.; Tanghe, A.; Denis, O.; Jurion, P.; Castiglione, N.; Vanonckelen, A.; Palfliet, K.; Huygen, K. Mapping of murine Th1 helper T-cell epitopes of mycolyl transferases Ag85A, Ag85B, and Ag85C from Mycobacterium tuberculosis. Infect. Immun. 2003, 71, 483–493. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ramírez-Gualito, K.; Richter, M.; Matzapetakis, M.; Singer, D.; Berger, S. Structural Characterization by NMR of a Double Phosphorylated Chimeric Peptide Vaccine for Treatment of Alzheimer’s Disease. Molecules 2013, 18, 4929-4941. https://doi.org/10.3390/molecules18054929
Ramírez-Gualito K, Richter M, Matzapetakis M, Singer D, Berger S. Structural Characterization by NMR of a Double Phosphorylated Chimeric Peptide Vaccine for Treatment of Alzheimer’s Disease. Molecules. 2013; 18(5):4929-4941. https://doi.org/10.3390/molecules18054929
Chicago/Turabian StyleRamírez-Gualito, Karla, Monique Richter, Manolis Matzapetakis, David Singer, and Stefan Berger. 2013. "Structural Characterization by NMR of a Double Phosphorylated Chimeric Peptide Vaccine for Treatment of Alzheimer’s Disease" Molecules 18, no. 5: 4929-4941. https://doi.org/10.3390/molecules18054929
APA StyleRamírez-Gualito, K., Richter, M., Matzapetakis, M., Singer, D., & Berger, S. (2013). Structural Characterization by NMR of a Double Phosphorylated Chimeric Peptide Vaccine for Treatment of Alzheimer’s Disease. Molecules, 18(5), 4929-4941. https://doi.org/10.3390/molecules18054929