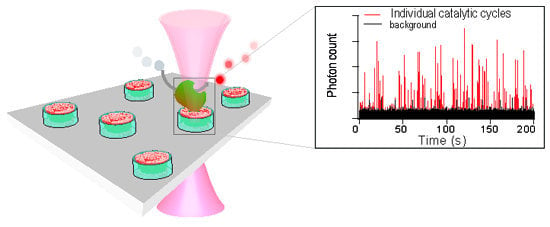
Shedding Light on Protein Folding, Structural and Functional Dynamics by Single Molecule Studies
Abstract
:1. Introduction
2. Single Molecule Insights into Protein Structural and Functional Dynamics
2.1. Methods to Directly Observe Protein Dynamics of Protein Folding and Conformational Sampling

2.1.1. Single Molecule Insights into Protein Folding Mechanisms

2.1.2. Single Molecule Insights into the Role of Protein Conformational Dynamics to Function

2.2. Methods to Attain Single Molecule Insights in Protein Functional Dynamics

2.2.1. Dynamic Disorder: Sampling of a Few or a Spectrum of Activity States?

2.2.2. Static Disorder: Multiple Distinct Folds of the Same Sequence or Chemical Heterogeneities?

3. Current Improvements and Future Directions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Michalet, X.; Weiss, S.; Jager, M. Single-molecule fluorescence studies of protein folding and conformational dynamics. Chem. Rev. 2006, 106, 1785–1813. [Google Scholar] [PubMed]
- Jorgensen, S.K.; Hatzakis, N.S. Insights in enzyme functional dynamics and activity regulation by single molecule studies. Biophys. Rev. Lett. 2013, 8, 137–160. [Google Scholar] [CrossRef]
- Moerner, W.E. New directions in single-molecule imaging and analysis. Proc. Natl. Acad. Sci. USA 2007, 104, 12596–12602. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.S.; Choi, P.J.; Li, G.W.; Lee, N.K.; Lia, G. Single-molecule approach to molecular biology in living bacterial cells. Annu. Rev. Biophys. 2008, 37, 417–444. [Google Scholar] [CrossRef] [PubMed]
- Schuler, B.; Eaton, W.A. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 2008, 18, 16–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joo, C.; Balci, H.; Ishitsuka, Y.; Buranachai, C.; Ha, T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008, 77, 51–76. [Google Scholar] [CrossRef] [PubMed]
- Cornish, P.V.; Ha, T. A survey of single-molecule techniques in chemical biology. ACS Chem. Biol. 2007, 2, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.R.; Deniz, A.A. Shedding light on protein folding landscapes by single-molecule fluorescence. Chem. Soc. Rev. 2014, 43, 1172–1188. [Google Scholar] [CrossRef] [PubMed]
- Hatzakis, N.S. Single molecule insights on conformational selection and induced fit mechanism. Biophys. Chem. 2014, 186, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Luo, G.B.; Karnchanaphanurach, P.; Louie, T.M.; Rech, I.; Cova, S.; Xun, L.Y.; Xie, X.S. Protein conformational dynamics probed by single-molecule electron transfer. Science 2003, 302, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.P.; Xun, L.Y.; Xie, X.S. Single-molecule enzymatic dynamics. Science 1998, 282, 1877–1882. [Google Scholar] [CrossRef] [PubMed]
- Guydosh, N.R.; Block, S.M. Direct observation of the binding state of the kinesin head to the microtubule. Nature 2009, 461, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.F.; Terry, D.S.; Shi, L.; Quick, M.; Weinstein, H.; Blanchard, S.C.; Javitch, J.A. Substrate-modulated gating dynamics in a Na(+)-coupled neurotransmitter transporter homologue. Nature 2011, 474, 109–113. [Google Scholar] [CrossRef]
- Mukhtasimova, N.; Lee, W.Y.; Wang, H.L.; Sine, S.M. Detection and trapping of intermediate states priming nicotinic receptor channel opening. Nature 2009, 459, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.J.; Cai, L.; Frieda, K.; Xie, S. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science 2008, 322, 442–446. [Google Scholar] [CrossRef]
- Laursen, T.; Singha, A.; Rantzau, N.; Tutkus, M.; Borch, J.; Hedegard, P.; Stamou, D.; Møller, B.L.; Hatzakis, N.S. Single Molecule Activity Measurements of Cytochrome P450 Oxidoreductase Reveal the Existence of Two Discrete Functional States. ACS Chem. Biol. 2014, 9, 630–634. [Google Scholar] [PubMed]
- Hatzakis, N.S.; Wei, L.; Jorgensen, S.K.; Kunding, A.H.; Bolinger, P.Y.; Ehrlich, N.; Makarov, I.; Skjot, M.; Svendsen, A.; Hedegard, P.; et al. Single enzyme studies reveal the existence of discrete functional states for monomeric enzymes and how they are “selected” upon allosteric regulation. J. Am. Chem. Soc. 2012, 134, 9296–9302. [Google Scholar] [CrossRef] [PubMed]
- Rissin, D.M.; Gorris, H.H.; Walt, D.R. Distinct and long-lived activity states of single enzyme molecules. J. Am. Chem. Soc. 2008, 130, 5349–5353. [Google Scholar] [CrossRef] [PubMed]
- Velonia, K.; Flomenbom, O.; Loos, D.; Masuo, S.; Cotlet, M.; Engelborghs, Y.; Hofkens, J.; Rowan, A.E.; Klafter, J.; Nolte, R.J.M.; et al. Single-enzyme kinetics of CALB-catalyzed hydrolysis. Angew. Chem. Int. Ed. 2005, 44, 560–564. [Google Scholar] [CrossRef]
- Iversen, L.; Tu, H.L.; Lin, W.C.; Christensen, S.M.; Abel, S.M.; Iwig, J.; Wu, H.J.; Gureasko, J.; Rhodes, C.; Petit, R.S.; et al. Ras activation by SOS: Allosteric regulation by altered fluctuation dynamics. Science 2014, 345, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; Kumar, S.; Ma, B.Y.; Nussinov, R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999, 8, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Teilum, K.; Olsen, J.G.; Kragelund, B.B. Functional aspects of protein flexibility. Cell. Mol. Life Sci. 2009, 66, 2231–2247. [Google Scholar] [CrossRef] [PubMed]
- Claessen, V.I.; Engelkamp, H.; Christianen, P.C.; Maan, J.C.; Nolte, R.J.; Blank, K.; Rowan, A.E. Single-biomolecule kinetics: the art of studying a single enzyme. Annu. Rev. Anal. Chem. 2010, 3, 319–340. [Google Scholar] [CrossRef]
- Walter, N.G.; Huang, C.Y.; Manzo, A.J.; Sobhy, M.A. Do-it-yourself guide: How to use the modern single-molecule toolkit. Nat. Methods 2008, 5, 475–489. [Google Scholar] [CrossRef] [PubMed]
- Hohng, S.; Lee, S.; Lee, J.; Jo, M.H. Maximizing information content of single-molecule FRET experiments: multi-color FRET and FRET combined with force or torque. Chem. Soc. Rev. 2014, 43, 1007–1013. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef]
- Oddershede, L.B. Force probing of individual molecules inside the living cell is now a reality. Nat. Chem. Biol. 2012, 8, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Greenleaf, W.J.; Woodside, M.T.; Block, S.M. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Fazal, F.M.; Block, S.M. Optical tweezers study life under tension. Nat. Photonics 2011, 5, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.; Muller, D.J. Observing single biomolecules at work with the atomic force microscope. Nat. Struct. Biol. 2000, 7, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.E.; Marszalek, P.E.; Fernandez, J.M. Stretching single molecules into novel conformations using the atomic force microscope. Nat. Struct. Biol. 2000, 7, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Dufrene, Y.F. Towards nanomicrobiology using atomic force microscopy. Nat. Rev. Microbiol. 2008, 6, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.J.; Dufrene, Y.F. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat. Nanotech. 2008, 3, 261–269. [Google Scholar] [CrossRef]
- Muller, D.J.; Helenius, J.; Alsteens, D.; Dufrene, Y.F. Force probing surfaces of living cells to molecular resolution. Nat. Chem. Biol. 2009, 5, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Pinaud, F.; Clarke, S.; Sittner, A.; Dahan, M. Probing cellular events, one quantum dot at a time. Nat. Methods 2010, 7, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Yang, H. Progress in single-molecule spectroscopy in cells. Curr. Opin. Chem. Biol. 2010, 14, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Veigel, C.; Schmidt, C.F. Moving into the cell: Single-molecule studies of molecular motors in complex environments. Nat. Rev. Mol. Cell Bio. 2011, 12, 163–176. [Google Scholar] [CrossRef]
- Ha, T.; Kozlov, A.G.; Lohman, T.M. Single-molecule views of protein movement on single-stranded DNA. Annu. Rev. Biophys. 2012, 41, 295–319. [Google Scholar] [CrossRef] [PubMed]
- Engelkamp, H.; Hatzakis, N.S.; Hofkens, J.; de Schryver, F.C.; Nolte, R.J.M.; Rowan, A.E. Do enzymes sleep and work? Chem. Commun. 2006, 935–940. [Google Scholar]
- Smiley, R.D.; Hammes, G.G. Single molecule studies of enzyme mechanisms. Chem. Rev. 2006, 106, 3080–3094. [Google Scholar] [CrossRef]
- Min, W.; English, B.P.; Luo, G.B.; Cherayil, B.J.; Kou, S.C.; Xie, X.S. Fluctuating enzymes: Lessons from single-molecule studies. Acc. Chem. Res. 2005, 38, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Tinnefeld, P.; Sauer, M. Branching out of single-molecule fluorescence spectroscopy: Challenges for chemistry and influence on biology. Angew. Chem. Int. Ed. 2005, 44, 2642–2671. [Google Scholar] [CrossRef]
- Roeffaers, M.B.J.; de Cremer, G.; Uji-i, H.; Muls, B.; Sels, B.F.; Jacobs, P.A.; de Schryver, F.C.; de Vos, D.E.; Hofkens, J. Single-molecule fluorescence spectroscopy in (bio)catalysis. Proc. Natl. Acad. Sci. USA 2007, 104, 12603–12609. [Google Scholar] [CrossRef] [PubMed]
- Hohlbein, J.; Aigrain, L.; Craggs, T.D.; Bermek, O.; Potapova, O.; Shoolizadeh, P.; Grindley, N.D.F.; Joyce, C.M.; Kapanidis, A.N. Conformational landscapes of DNA polymerase I and mutator derivatives establish fidelity checkpoints for nucleotide insertion. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Chung, H.S.; Eaton, W.A. Single-molecule fluorescence probes dynamics of barrier crossing. Nature 2013, 502, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Soranno, A.; Koenig, I.; Borgia, M.B.; Hofmann, H.; Zosel, F.; Nettels, D.; Schuler, B. Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments. Proc. Natl. Acad. Sci. USA 2014, 111, 4874–4879. [Google Scholar] [CrossRef] [PubMed]
- Cisse, I.; Okumus, B.; Joo, C.; Ha, T. Fueling protein–DNA interactions inside porous nanocontainers. Proc. Natl. Acad. Sci. USA 2007, 104, 12646–12650. [Google Scholar] [CrossRef] [PubMed]
- Stryer, L.; Haugland, R.P. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. USA 1967, 58, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.B.; Kunzelmann, S.; Webb, M.R.; Ha, T. Detecting intramolecular conformational dynamics of single molecules in short distance range with subnanometer sensitivity. Nano Lett. 2011, 11, 5482–5488. [Google Scholar] [CrossRef] [PubMed]
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Bryngelson, J.D.; Wolynes, P.G. Spin-glasses and the statistical-mechanics of protein folding. Proc. Natl. Acad. Sci. USA 1987, 84, 7524–7528. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M. Behind the folding funnel diagram. Nat. Chem. Biol. 2011, 7, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Onuchic, J.N.; Luthey-Schulten, Z.; Wolynes, P.G. Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 1997, 48, 545–600. [Google Scholar] [CrossRef] [PubMed]
- Tanford, C. Protein denaturation. Adv. Protein Chem. 1968, 23, 121–282. [Google Scholar] [PubMed]
- Kim, P.S.; Baldwin, R.L. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu. Rev. Biochem. 1982, 51, 459–489. [Google Scholar] [CrossRef] [PubMed]
- Frauenfelder, H.; Sligar, S.G.; Wolynes, P.G. The energy landscapes and motions of proteins. Science 1991, 254, 1598–1603. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.; Koga, N.; Takada, S.; Onuchic, J.N.; Wolynes, P.G. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2006, 103, 11844–11849. [Google Scholar] [CrossRef] [PubMed]
- Bryngelson, J.D.; Onuchic, J.N.; Socci, N.D.; Wolynes, P.G. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins 1995, 21, 167–195. [Google Scholar] [PubMed]
- Dill, K.A.; Chan, H.S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997, 4, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.P. Sizing up single-molecule enzymatic conformational dynamics. Chem. Soc. Rev. 2014, 43, 1118–1143. [Google Scholar] [CrossRef] [PubMed]
- Schuler, B.; Hofmann, H. Single-molecule spectroscopy of protein folding dynamics—Expanding scope and timescales. Curr. Opin. Struct. Biol. 2013, 23, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Chattopadhyay, K. Studies of protein folding and dynamics using single molecule fluorescence spectroscopy. Phys. Chem. Chem. Phys. 2014, 16, 11139–11149. [Google Scholar] [CrossRef] [PubMed]
- Borgia, A.; Williams, P.M.; Clarke, J. Single-molecule studies of protein folding. Annu. Rev. Biochem. 2008, 77, 101–125. [Google Scholar] [CrossRef]
- Deniz, A.A.; Laurence, T.A.; Beligere, G.S.; Dahan, M.; Martin, A.B.; Chemla, D.S.; Dawson, P.E.; Schultz, P.G.; Weiss, S. Single-molecule protein folding: Diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc. Natl. Acad. Sci. USA 2000, 97, 5179–5184. [Google Scholar] [CrossRef] [PubMed]
- Schuler, B.; Lipman, E.A.; Eaton, W.A. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature 2002, 419, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Groll, J.; Amirgoulova, E.V.; Ameringer, T.; Heyes, C.D.; Rocker, C.; Nienhaus, G.U.; Moller, M. Biofunctionalized, ultrathin coatings of cross-linked star-shaped poly(ethylene oxide) allow reversible folding of immobilized proteins. J. Am. Chem. Soc. 2004, 126, 4234–4239. [Google Scholar] [CrossRef] [PubMed]
- Campos, L.A.; Sadqi, M.; Liu, J.; Wang, X.; English, D.S.; Muñoz, V. Gradual disordering of the native state on a slow two-state folding protein monitored by single-molecule fluorescence spectroscopy and NMR. J. Phys. Chem. B 2013, 117, 13120–13131. [Google Scholar] [CrossRef] [PubMed]
- Lipman, E.A.; Schuler, B.; Bakajin, O.; Eaton, W.A. Single-molecule measurement of protein folding kinetics. Science 2003, 301, 1233–1235. [Google Scholar] [CrossRef] [PubMed]
- Kauffmann, E.; Darnton, N.C.; Austin, R.H.; Batt, C.; Gerwert, K. Lifetimes of intermediates in the beta -sheet to alpha-helix transition of beta-lactoglobulin by using a diffusional IR mixer. Proc. Natl. Acad. Sci. USA 2001, 98, 6646–6649. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, B.; Nettels, D.; Benke, S.; Clark, J.; Weidner, S.; Hofmann, H.; Pfeil, S.H.; Schuler, B. Microfluidic mixer designed for performing single-molecule kinetics with confocal detection on timescales from milliseconds to minutes. Nat. Protoc. 2013, 8, 1459–1474. [Google Scholar] [CrossRef] [PubMed]
- Talaga, D.S.; Lau, W.L.; Roder, H.; Tang, J.Y.; Jia, Y.W.; DeGrado, W.F.; Hochstrasser, R.M. Dynamics and folding of single two-stranded coiled-coil peptides studied by fluorescent energy transfer confocal microscopy. Proc. Natl. Acad. Sci. USA 2000, 97, 13021–13026. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Palfey, B.A.; Dertouzos, J.; Jensen, K.F.; Gafni, A.; Steel, D. Multiple states of the Tyr318Leu mutant of dihydroorotate dehydrogenase revealed by single-molecule kinetics. J. Am. Chem. Soc. 2004, 126, 6914–6922. [Google Scholar] [CrossRef]
- Rhoades, E.; Gussakovsky, E.; Haran, G. Watching proteins fold one molecule at a time. Proc. Natl. Acad. Sci. USA 2003, 100, 3197–3202. [Google Scholar] [CrossRef] [PubMed]
- Pirchi, M.; Ziv, G.; Riven, I.; Cohen, S.S.; Zohar, N.; Barak, Y.; Haran, G. Single-molecule fluorescence spectroscopy maps the folding landscape of a large protein. Nat. Commun. 2011, 2, 493. [Google Scholar] [CrossRef] [PubMed]
- Panchenko, A.R.; LutheySchulten, Z.; Cole, R.; Wolynes, P.G. The foldon universe: A survey of structural similarity and self-recognition of independently folding units. J. Mol. Biol. 1997, 272, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; McHale, K.; Louis, J.M.; Eaton, W.A. Single-molecule fluorescence experiments determine protein folding transition path times. Science 2012, 335, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.M.; van der Lee, R.; de Groot, N.S.; Gsponer, J. Intrinsically disordered proteins: Regulation and disease. Curr. Opin. Struct. Biol. 2011, 21, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 2008, 37, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graphics Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Krishnan, R.; Lemke, E.A.; Lindquist, S.; Deniz, A.A. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl. Acad. Sci. USA 2007, 104, 2649–2654. [Google Scholar] [CrossRef] [PubMed]
- Ferreon, A.C.; Gambin, Y.; Lemke, E.A.; Deniz, A.A. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA 2009, 106, 5645–5650. [Google Scholar] [CrossRef] [PubMed]
- Ferreon, A.C.; Moran, C.R.; Ferreon, J.C.; Deniz, A.A. Alteration of the alpha-synuclein folding landscape by a mutation related to Parkinson’s disease. Angew. Chem. Int. Ed. 2010, 49, 3469–3472. [Google Scholar] [CrossRef]
- Sevcsik, E.; Trexler, A.J.; Dunn, J.M.; Rhoades, E. Allostery in a disordered protein: Oxidative modifications to alpha-synuclein act distally to regulate membrane binding. J. Am. Chem. Soc. 2011, 133, 7152–7158. [Google Scholar] [CrossRef] [PubMed]
- Trexler, A.J.; Rhoades, E. Single molecule characterization of alpha-synuclein in aggregation-prone states. Biophys. J. 2010, 99, 3048–3055. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.; Miranker, A.D.; Rhoades, E. A membrane-bound antiparallel dimer of rat islet amyloid polypeptide. Angew. Chem. Int. Ed. 2011, 50, 10859–10862. [Google Scholar] [CrossRef]
- Elbaum-Garfinkle, S.; Rhoades, E. Identification of an aggregation-prone structure of tau. J. Am. Chem. Soc. 2012, 134, 16607–16613. [Google Scholar] [CrossRef] [PubMed]
- Ferreon, A.C.M.; Ferreon, J.C.; Wright, P.E.; Deniz, A.A. Modulation of allostery by protein intrinsic disorder. Nature 2013, 498, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.; Nettels, D.; Schuler, B. Single-molecule spectroscopy of the unexpected collapse of an unfolded protein at low pH. J. Chem. Phys. 2013, 139, 121930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wuttke, R.; Hofmann, H.; Nettels, D.; Borgia, M.B.; Mittal, J.; Best, R.B.; Schuler, B. Temperature-dependent solvation modulates the dimensions of disordered proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 5213–5218. [Google Scholar] [CrossRef] [PubMed]
- Brucale, M.; Schuler, B.; Samori, B. Single-molecule studies of intrinsically disordered proteins. Chem. Rev. 2014, 114, 3281–3317. [Google Scholar] [CrossRef] [PubMed]
- Henzler-Wildman, K.; Kern, D. Dynamic personalities of proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Boehr, D.D.; Nussinov, R.; Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009, 5, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Mccammon, J.A.; Gelin, B.R.; Karplus, M. Dynamics of folded proteins. Nature 1977, 267, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Glowacki, D.R.; Harvey, J.N.; Mulholland, A.J. Taking Ockham’s razor to enzyme dynamics and catalysis. Nat. Chem. 2012, 4, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Bhabha, G.; Lee, J.; Ekiert, D.C.; Gam, J.; Wilson, I.A.; Dyson, H.J.; Benkovic, S.J.; Wright, P.E. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science 2011, 332, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Goodey, N.M.; Benkovic, S.J. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 2008, 4, 474–482. [Google Scholar] [CrossRef]
- Tzeng, S.R.; Kalodimos, C.G. Dynamic activation of an allosteric regulatory protein. Nature 2009, 462, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Kamerlin, S.C.L.; Warshel, A. At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis? Proteins-Struct. Funct. Bioinf. 2010, 78, 1339–1375. [Google Scholar]
- Henzler-Wildman, K.A.; Thai, V.; Lei, M.; Ott, M.; Wolf-Watz, M.; Fenn, T.; Pozharski, E.; Wilson, M.A.; Petsko, G.A.; Karplus, M.; et al. Intrinsic motions along an enzymatic reaction trajectory. Nature 2007, 450, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Mickler, M.; Hessling, M.; Ratzke, C.; Buchner, J.; Hugel, T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat. Struct. Mol. Bio. 2009, 16, 281–286. [Google Scholar] [CrossRef]
- Pisliakov, A.V.; Cao, J.; Kamerlin, S.C.; Warshel, A. Enzyme millisecond conformational dynamics do not catalyze the chemical step. Proc. Natl. Acad. Sci. USA 2009, 106, 17359–17364. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Lee, S.; Jeon, A.; Choi, J.M.; Lee, H.S.; Hohng, S.; Kim, H.S. A single-molecule dissection of ligand binding to a protein with intrinsic dynamics. Nat. Chem. Biol. 2013, 9, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.H.; Park, J.; Kim, E.; Hohng, S.; Kim, H.S. Protein conformational dynamics dictate the binding affinity for a ligand. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Rotman, B. Measurement of activity of single molecules of beta-d-galactosidase. Proc. Natl. Acad. Sci. USA 1961, 47, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- DeWitt, M.A.; Chang, A.Y.; Combs, P.A.; Yildiz, A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science 2012, 335, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Rocha, S.; Hutchison, J.A.; Peneva, K.; Herrmann, A.; Muellen, K.; Skjot, M.; Jorgensen, C.I.; Svendsen, A.; de Schryver, F.C.; Hofkens, J.; et al. Linking phospholipase mobility to activity by single-molecule wide-field microscopy. ChemPhysChem 2009, 10, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Gudmand, M.; Rocha, S.; Hatzakis, N.S.; Peneva, K.; Mullen, K.; Stamou, D.; Uji-I, H.; Hofkens, J.; Bjornholm, T.; Heimburg, T. Influence of lipid heterogeneity and phase behavior on phospholipase A(2) action at the single molecule level. Biophys. J. 2010, 98, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Brender, J.R.; Dertouzos, J.; Ballou, D.P.; Massey, V.; Palfey, B.A.; Entsch, B.; Steel, D.G.; Gafni, A. Conformational dynamics of the isoalloxazine in substrate-free p-hydroxybenzoate hydroxylase: Single-molecule studies. J. Am. Chem. Soc. 2005, 127, 18171–18178. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Dertouzos, J.; Gafni, A.; Steel, D.; Palfey, B.A. Single-molecule kinetics reveals signatures of half-sites reactivity in dihydroorotate dehydrogenase A catalysis. Proc. Natl. Acad. Sci. USA 2006, 103, 5775–5780. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, R.H.; Tabares, L.C.; Kostrz, D.; Dennison, C.; Aartsma, T.J.; Canters, G.W.; Moerner, W.E. Redox cycling and kinetic analysis of single molecules of solution-phase nitrite reductase. Proc. Natl. Acad. Sci. USA 2011, 108, 17269–17274. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, S.; Zauner, G.; Aartsma, T.J.; Engelkamp, H.; Hatzakis, N.; Rowan, A.E.; Nolte, R.J.M.; Christianen, P.C.M.; Canters, G.W. The enzyme mechanism of nitrite reductase studied at single-molecule level. Proc. Natl. Acad. Sci. USA 2008, 105, 3250–3255. [Google Scholar] [CrossRef]
- English, B.P.; Min, W.; van Oijen, A.M.; Lee, K.T.; Luo, G.; Sun, H.; Cherayil, B.J.; Kou, S.C.; Xie, X.S. Ever-fluctuating single enzyme molecules: Michaelis-Menten equation revisited. Nat. Chem. Biol. 2006, 2, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Hatzakis, N.S.; Engelkamp, H.; Velonia, K.; Hofkens, J.; Christianen, P.C.M.; Svendsen, A.; Patkar, S.A.; Vind, J.; Maan, J.C.; Rowan, A.E.; et al. Synthesis and single enzyme activity of a clicked lipase-BSA hetero-dimer. Chem. Commun. 2006, 2012–2014. [Google Scholar]
- De Cremer, G.; Roeffaers, M.B.J.; Baruah, M.; Sliwa, M.; Sels, B.F.; Hofkens, J.; de Vos, D.E. Dynamic disorder and stepwise deactivation in a chymotrypsin catalyzed hydrolysis reaction. J. Am. Chem. Soc. 2007, 129, 15458–15459. [Google Scholar] [CrossRef] [PubMed]
- Edman, L.; Foldes-Papp, Z.; Wennmalm, S.; Rigler, R. The fluctuating enzyme: A single molecule approach. Chem. Phys. 1999, 247, 11–22. [Google Scholar] [CrossRef]
- Xie, X.S.; Lu, H.P. Single-molecule Enzymology. J. Biol. Chem. 1999, 274, 15967–15970. [Google Scholar] [CrossRef] [PubMed]
- Veshaguri, S.; Tutkus, M.; Lundgaard, C.V.; Tonnesen, A.; Mathiasen, S.; Jorgensen, S.K.; Iversen, L.; Hatzakis, N.S.; Rasmussen, S.G.; Stamou, D. Developing an assay to probe activation and conformational dynamics of β2-adrenergic receptor on single molecule level. Biophys. J. 2013, 104. [Google Scholar] [CrossRef]
- Elizondo, E.; Larsen, J.; Hatzakis, N.S.; Cabrera, I.; Bjornhorn, T.; Veciana, J.; Stamou, D.; Ventosa, N. Influence of the Preparation Route on the Supramolecular Organization of Lipids in a Vesicular System. J. Am. Chem. Soc. 2012, 134, 1918–1921. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.M.; Bolinger, P.Y.; Hatzakis, N.S.; Mortensen, M.W.; Stamou, D. Mixing subattolitre volumes in a quantitative and highly parallel manner with soft matter nanofluidics. Nat. Nanotech. 2012, 7, 51–55. [Google Scholar] [CrossRef]
- Larsen, J.; Hatzakis, N.S.; Stamou, D. Observation of inhomogeneity in the lipid composition of individual nanoscale liposomes. J. Am. Chem. Soc. 2011, 133, 10685–10687. [Google Scholar] [CrossRef] [PubMed]
- Hatzakis, N.S.; Bhatia, V.K.; Larsen, J.; Madsen, K.L.; Bolinger, P.Y.; Kunding, A.H.; Castillo, J.; Gether, U.; Hedegard, P.; Stamou, D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol. 2009, 5, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.K.; Hatzakis, N.S.; Stamou, D. A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Semin. Cell Dev. Biol. 2010, 21, 381–390. [Google Scholar] [PubMed]
- Lohr, C.; Kunding, A.H.; Bhatia, V.K.; Stamou, D. Constructing size distributions of liposomes from single-object fluorescence measurements. Methods Enzymol. 2009, 465, 143–160. [Google Scholar] [PubMed]
- Bhatia, V.K.; Madsen, K.L.; Bolinger, P.Y.; Kunding, A.; Hedegard, P.; Gether, U.; Stamou, D. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009, 28, 3303–3314. [Google Scholar] [CrossRef] [PubMed]
- Bendix, P.M.; Pedersen, M.S.; Stamou, D. Quantification of nano-scale intermembrane contact areas by using fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2009, 106, 12341–12346. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. Laboratory evolution of stereoselective enzymes: A prolific source of catalysts for asymmetric reactions. Angew. Chem. Int. Ed. 2011, 50, 138–174. [Google Scholar] [CrossRef]
- Hatzakis, N.S.; Daphnomili, D.; Smonou, I. Ferulic acid esterase from Humicola Insolens catalyzes enantioselective transesterification of secondary alcohols. J. Mol. Catal. B: Enzym. 2003, 21, 309–311. [Google Scholar] [CrossRef]
- Hatzakis, N.S.; Smonou, I. Asymmetric transesterification of secondary alcohols catalyzed by feruloyl esterase from Humicola insolens. Bioorg. Chem. 2005, 33, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.D.; Verger, R. Lipases: Interfacial enzymes with attractive applications. Angew. Chem. Int. Ed. 1998, 37, 1609–1633. [Google Scholar] [CrossRef]
- Hatzakis, N.S.; Smonou, I. Enantioselectivity and diastereoselectivity in the transesterification of secondary alcohols mediated by feruloyl esterase from Humicola insolens. Tetrahedron Lett. 2004, 45, 2755–2757. [Google Scholar] [CrossRef]
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef]
- Kazlauskas, R.J.; Bornscheuer, U.T. Finding better protein engineering strategies. Nat. Chem. Biol. 2009, 5, 526–529. [Google Scholar] [CrossRef] [PubMed]
- Cajal, Y.; Berg, O.G.; Jain, M.K. Origins of delays in monolayer kinetics: Phospholipase A(2) paradigm. Biochemistry 2004, 43, 9256–9264. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Sligar, S.G. Membrane protein assembly into nanodiscs. FEBS Lett. 2010, 584, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Laursen, T.; Jensen, K.; Møller, B.L. Conformational changes of the NADPH-dependent cytochrome P450 reductase in the course of electron transfer to cytochromes P450. Biochim. Biophys. Acta 2011, 1814, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.; Møller, B.L. Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry 2010, 71, 132–141. [Google Scholar] [PubMed]
- Fluck, C.E.; Tajima, T.; Pandey, A.V.; Arlt, W.; Okuhara, K.; Verge, C.F.; Jabs, E.W.; Mendonca, B.B.; Fujieda, K.; Miller, W.L. Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat. Genet. 2004, 36, 228–230. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Agrawal, V.; Giacomini, K.M.; Miller, W.L. Genetics of P450 oxidoreductase: Sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc. Natl. Acad. Sci. USA 2008, 105, 1733–1738. [Google Scholar]
- Xia, C.W.; Panda, S.P.; Marohnic, C.C.; Martasek, P.; Masters, B.S.; Kim, J.J.P. Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency. Proc. Natl. Acad. Sci. USA 2011, 108, 13486–13491. [Google Scholar] [CrossRef] [PubMed]
- Li, C.B.; Yang, H.; Kornatsuzaki, T. Multiscale complex network of protein conformational fluctuations in single-molecule time series. Proc. Natl. Acad. Sci. USA 2008, 105, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.M.; Craik, C.S. Trapping moving targets with small molecules. Science 2009, 324, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Boehr, D.D.; Dyson, H.J.; Wright, P.E. An NMR perspective on enzyme dynamics. Chem. Rev. 2006, 106, 3055–3079. [Google Scholar] [CrossRef] [PubMed]
- Smock, R.G.; Gierasch, L.M. Sending Signals Dynamically. Science 2009, 324, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, P.T.R.; Zhang, Z.Q.; McCourt, L.; Dwyer, M.; Benkovic, S.J.; Hammes, G.G. Interaction of dihydrofolate reductase with methotrexate: Ensemble and single-molecule kinetics. Proc. Natl. Acad. Sci. USA 2002, 99, 13481–13486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Rajagopalan, P.T.R.; Selzer, T.; Benkovic, S.J.; Hammes, G.G. Single-molecule and transient kinetics investigation of the interaction of dihydrofolate reductase with NADPH and dihydrofolate. Proc. Natl. Acad. Sci. USA 2004, 101, 2764–2769. [Google Scholar] [CrossRef] [PubMed]
- Lerch, H.P.; Rigler, R.; Mikhailov, A.S. Functional conformational motions in the turnover cycle of cholesterol oxidase. Proc. Natl. Acad. Sci. USA 2005, 102, 10807–10812. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Groote, R.; Schonherr, H.; Vancso, G.J. Probing single enzyme kinetics in real-time. Chem. Soc. Rev. 2009, 38, 2671–2683. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Moody, I.S.; Sims, P.C.; Hunt, S.R.; Corso, B.L.; Perez, I.; Weiss, G.A.; Collins, P.G. Single-Molecule Lysozyme Dynamics Monitored by an Electronic Circuit. Science 2012, 335, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Lerch, H.P.; Mikhailov, A.S.; Hess, B. Conformational-relaxation models of single-enzyme kinetics. Proc. Natl. Acad. Sci. USA 2002, 99, 15410–15415. [Google Scholar]
- Terentyeva, T.G.; Engelkamp, H.; Rowan, A.E.; Komatsuzaki, T.; Hofkens, J.; Li, C.B.; Blank, K. Dynamic disorder in single-enzyme experiments: Facts and artifacts. ACS Nano 2012, 6, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Terentyeva, T.G.; Hofkens, J.; Komatsuzaki, T.; Blank, K.; Li, C.B. Time-resolved single molecule fluorescence spectroscopy of an alpha-Chymotrypsin catalyzed reaction. J. Phys. Chem. B 2013, 117, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Moody, I.S.; Sims, P.C.; Hunt, S.R.; Corso, B.L.; Seitz, D.E.; Blaszcazk, L.C.; Collins, P.G.; Weiss, G.A. Single-molecule dynamics of lysozyme processing distinguishes linear and cross-linked peptidoglycan substrates. J. Am. Chem. Soc. 2012, 134, 2032–2035. [Google Scholar] [CrossRef] [PubMed]
- Flomenbom, O.; Silbey, R.J. Toolbox for analyzing finite two-state trajectories. Phys. Rev. E 2008, 78. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.Y.; Nussinov, R. Enzyme dynamics point to stepwise conformational selection in catalysis. Curr. Opin. Chem. Biol. 2010, 14, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Nagel, Z.D.; Klinman, J.P. A 21(st) century revisionist’s view at a turning point in enzymology. Nat. Chem. Biol. 2009, 5, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Lyon, A.M.; Tesmer, V.M.; Dhamsania, V.D.; Thal, D.M.; Gutierrez, J.; Chowdhury, S.; Suddala, K.C.; Northup, J.K.; Tesmer, J.J.G. An autoinhibitory helix in the C-terminal region of phospholipase C-beta mediates G alpha(q) activation. Nat. Struct. Mol. Biol. 2011, 18, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Volkman, B.F.; Lipson, D.; Wemmer, D.E.; Kern, D. Two-state allosteric behavior in a single-domain signaling protein. Science 2001, 291, 2429–2433. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.F.; Yeung, E.S. Differences in the chemical reactivity of individual molecules of an enzyme. Nature 1995, 373, 681–683. [Google Scholar] [CrossRef] [PubMed]
- Polakowski, R.; Craig, D.B.; Skelley, A.; Dovichi, N.J. Single molecules of highly purified bacterial alkaline phosphatase have identical activity. J. Am. Chem. Soc. 2000, 122, 4853–4855. [Google Scholar] [CrossRef]
- Gorris, H.H.; Rissin, D.M.; Walt, D.R. Stochastic inhibitor release and binding from single-enzyme molecules. Proc. Natl. Acad. Sci. USA 2007, 104, 17680–17685. [Google Scholar] [CrossRef] [PubMed]
- Rojek, M.J.; Walt, D.R. Observing single enzyme molecules interconvert between activity states upon heating. PLoS One 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Solomatin, S.V.; Greenfeld, M.; Chu, S.; Herschlag, D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature 2010, 463, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.A.; Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009, 10, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Liebherr, R.B.; Renner, M.; Gorris, H.H. A Single molecule perspective on the functional diversity of in vitro evolved beta-Glucuronidase. J. Am. Chem. Soc. 2014, 136, 5949–5955. [Google Scholar]
- Tokuriki, N.; Tawfik, D.S. Protein dynamism and evolvability. Science 2009, 324, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Piwonski, H.M.; Goomanovsky, M.; Bensimon, D.; Horovitz, A.; Haran, G. Allosteric inhibition of individual enzyme molecules trapped in lipid vesicles. Proc. Natl. Acad. Sci. USA 2012, 109, E1437–E1443. [Google Scholar] [CrossRef] [PubMed]
- Wadsater, M.; Laursen, T.; Singha, A.; Hatzakis, N.S.; Stamou, D.; Barker, R.; Mortensen, K.; Feidenhans’l, R.; Møller, B.L.; Cardenas, M. Monitoring shifts in the conformation equilibrium of the membrane protein cytochrome P450 reductase (POR) in nanodiscs. J. Biol. Chem. 2012, 287, 34596–34603. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.; Trexler, A.J.; Koo, P.; Miranker, A.D.; Atkins, W.M.; Rhoades, E. Single-molecule fluorescence spectroscopy using phospholipid bilayer nanodiscs. Method. Enzymol. 2010, 472, 89–117. [Google Scholar]
- Vriezema, D.M.; Garcia, P.M.L.; Oltra, N.S.; Hatzakis, N.S.; Kuiper, S.M.; Nolte, R.J.M.; Rowan, A.E.; van Hest, J.C.M. Positional assembly of enzymes in polymersome nanoreactors for cascade reactions. Angew. Chem. Int. Ed. 2007, 46, 7378–7382. [Google Scholar] [CrossRef]
- Felici, M.; Marza-Perez, M.; Hatzakis, N.S.; Nolte, R.J.M.; Feiters, M.C. beta-Cyclodextrin-appended giant amphiphile: Aggregation to vesicle polymersomes and immobilisation of enzymes. Chem. Eur. J. 2008, 14, 9914–9920. [Google Scholar] [CrossRef] [PubMed]
- Dirks, A.J.T.; van Berkel, S.S.; Hatzakis, N.S.; Opsteen, J.A.; van Delft, F.L.; Cornelissen, J.; Rowan, A.E.; van Hest, J.C.M.; Rutjes, F.; Nolte, R.J.M. Preparation of biohybrid amphiphiles via the copper catalysed Huisgen 3+2 dipolar cycloaddition reaction. Chem. Commun. 2005, 4172–4174. [Google Scholar]
- Discher, B.M.; Won, Y.Y.; Ege, D.S.; Lee, J.C.; Bates, F.S.; Discher, D.E.; Hammer, D.A. Polymersomes: Tough vesicles made from diblock copolymers. Science 1999, 284, 1143–1146. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, S.K.; McMillan, D.; Krzeminski, L.; Stamou, D.; Jeuken, L.J.C.; Hatzakis, N.S. Single proton pump activity measurements on single vesicles for a quinol heme-copper oxidase. Biophys. J. 2013, 104, 277a–278a. [Google Scholar] [CrossRef]
- Jørgensen, S.K.; Lassen, L.M.M.; Kemmer, G.; Gunther-Pomorski, T.; Stamou, D.; Bjørnholm, T.; Jensen, P.E.; Hatzakis, N.S. Creating a proteoliposome assay for single photosystem I activity assessment. Biophys. J. 2012, 102, 626a–627a. [Google Scholar] [CrossRef]
- Jain, A.; Liu, R.; Ramani, B.; Arauz, E.; Ishitsuka, Y.; Ragunathan, K.; Park, J.; Chen, J.; Xiang, Y.K.; Ha, T. Probing cellular protein complexes using single-molecule pull-down. Nature 2011, 473, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Yeom, K.H.; Heo, I.; Lee, J.; Hohng, S.; Kim, V.N.; Joo, C. Single-molecule approach to immunoprecipitated protein complexes: Insights into miRNA uridylation. EMBO Rep. 2011, 12, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Loveland, A.B.; Habuchi, S.; Walter, J.C.; van Oijen, A.M. A general approach to break the concentration barrier in single-molecule imaging. Nat. Methods 2012, 9, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Lang, K.; Chin, J.W. Bioorthogonal reactions for labeling proteins. ACS Chem. Biol. 2014, 9, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Moth-Poulsen, K.; Kofod-Hansen, V.; Kamounah, F.S.; Hatzakis, N.S.; Stamou, D.; Schaumburg, K.; Christensen, J.B. Optically induced linking of protein and nanoparticles to gold surfaces. Bioconjug. Chem. 2010, 21, 1056–1061. [Google Scholar] [CrossRef]
- Brennan, J.L.; Hatzakis, N.S.; Tshikhudo, T.R.; Dirvianskyte, N.; Razumas, V.; Patkar, S.; Vind, J.; Svendsen, A.; Nolte, R.J.M.; Rowan, A.E.; et al. Bionanoconjugation via click chemistry: The creation of functional hybrids of lipases and gold nanoparticles. Bioconjug. Chem. 2006, 17, 1373–1375. [Google Scholar] [CrossRef]
- Tornøe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on solid phase: [1,2,3]-Triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef] [PubMed]
- Milles, S.; Tyagi, S.; Banterle, N.; Koehler, C.; VanDelinder, V.; Plass, T.; Neal, A.P.; Lemke, E.A. Click strategies for single-molecule protein fluorescence. J. Am. Chem. Soc. 2012, 134, 5187–5195. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.; Davis, L.; Torres-Kolbus, J.; Chou, C.; Deiters, A.; Chin, J.W. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem. 2012, 4, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Uttamapinant, C.; Tangpeerachaikul, A.; Grecian, S.; Clarke, S.; Singh, U.; Slade, P.; Gee, K.R.; Ting, A.Y. Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew. Chem. Int. Ed. 2012, 51, 5852–5856. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, X.; Verhelst, S. Comparative analysis of click chemistry mediated activity-based protein profiling in cell lysates. Molecules 2013, 18, 12599–12608. [Google Scholar] [CrossRef] [PubMed]
- Rasnik, I.; McKinney, S.A.; Ha, T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods 2006, 3, 891–893. [Google Scholar] [CrossRef] [PubMed]
- Alejo, J.L.; Blanchard, S.C.; Andersen, O.S. Small-molecule photostabilizing agents are modifiers of lipid bilayer properties. Biophys. J. 2013, 104, 2410–2418. [Google Scholar] [CrossRef] [PubMed]
- Altman, R.B.; Terry, D.S.; Zhou, Z.; Zheng, Q.; Geggier, P.; Kolster, R.A.; Zhao, Y.; Javitch, J.A.; Warren, J.D.; Blanchard, S.C. Cyanine fluorophore derivatives with enhanced photostability. Nat. Methods 2012, 9, 68–71. [Google Scholar] [CrossRef]
- Tinnefeld, P.; Cordes, T. “Self-healing” dyes: Intramolecular stabilization of organic fluorophores. Nat. Methods 2012, 9, 426–427, author reply 427–428. [Google Scholar] [CrossRef] [PubMed]
- Altman, R.B.; Zheng, Q.; Zhou, Z.; Terry, D.S.; Warren, J.D.; Blanchard, S.C. Enhanced photostability of cyanine fluorophores across the visible spectrum. Nat. Methods 2012, 9, 428–429. [Google Scholar] [CrossRef] [PubMed]
- Bharill, S.; Chen, C.; Stevens, B.; Kaur, J.; Smilansky, Z.; Mandecki, W.; Gryczynski, I.; Gryczynski, Z.; Cooperman, B.S.; Goldman, Y.E. Enhancement of single-molecule fluorescence signals by colloidal silver nanoparticles in studies of protein translation. ACS Nano 2011, 5, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Escobedo, J.O.; Rusin, O.; Lim, S.; Strongin, R.M. NIR dyes for bioimaging applications. Curr. Opin. Chem. Biol. 2010, 14, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Kapanidis, A.N.; Lee, N.K.; Laurence, T.A.; Doose, S.; Margeat, E.; Weiss, S. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. USA 2004, 101, 8936–8941. [Google Scholar] [CrossRef] [PubMed]
- Kapanidis, A.N.; Laurence, T.A.; Lee, N.K.; Margeat, E.; Kong, X.X.; Weiss, S. Alternating-laser excitation of single molecules. Acc. Chem. Res. 2005, 38, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Hohng, S.; Joo, C.; Ha, T. Single-molecule three-color FRET. Biophys. J. 2004, 87, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, S.; Ragunathan, K.; Joo, C.; Ha, T.; Hohng, S. Single-molecule four-color FRET. Angew. Chem. Int. Ed. 2010, 49, 9922–9925. [Google Scholar] [CrossRef]
- Kalinin, S.; Peulen, T.; Sindbert, S.; Rothwell, P.J.; Berger, S.; Restle, T.; Goody, R.S.; Gohlke, H.; Seidel, C.A.M. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat. Methods 2012, 9, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Grashoff, C.; Hoffman, B.D.; Brenner, M.D.; Zhou, R.B.; Parsons, M.; Yang, M.T.; McLean, M.A.; Sligar, S.G.; Chen, C.S.; Ha, T.; et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 2010, 466, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Balci, H.; Jia, H.; Lohman, T.M.; Ha, T. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nat. Commun. 2013, 4, 1878. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xiao, J.; Ren, X.J.; Lao, K.Q.; Xie, X.S. Probing gene expression in live cells, one protein molecule at a time. Science 2006, 311, 1600–1603. [Google Scholar] [CrossRef] [PubMed]
- Wakamoto, Y.; Dhar, N.; Chait, R.; Schneider, K.; Signorino-Gelo, F.; Leibler, S.; McKinney, J.D. Dynamic Persistence of antibiotic-stressed mycobacteria. Science 2013, 339, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Pieraccini, S.; Saladino, G.; Cappelletti, G.; Cartelli, D.; Francescato, P.; Speranza, G.; Manitto, P.; Sironi, M. In silico design of tubulin-targeted antimitotic peptides. Nat. Chem. 2009, 1, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Teague, S.J. Implications of protein flexibility for drug discovery. Nat. Rev. Drug Discov. 2003, 2, 527–541. [Google Scholar] [PubMed]
- Rothlisberger, D.; Khersonsky, O.; Wollacott, A.M.; Jiang, L.; DeChancie, J.; Betker, J.; Gallaher, J.L.; Althoff, E.A.; Zanghellini, A.; Dym, O.; et al. Kemp elimination catalysts by computational enzyme design. Nature 2008, 453, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Althoff, E.A.; Clemente, F.R.; Doyle, L.; Rothlisberger, D.; Zanghellini, A.; Gallaher, J.L.; Betker, J.L.; Tanaka, F.; Barbas, C.F.; et al. De novo computational design of retro-aldol enzymes. Science 2008, 319, 1387–1391. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bavishi, K.; Hatzakis, N.S. Shedding Light on Protein Folding, Structural and Functional Dynamics by Single Molecule Studies. Molecules 2014, 19, 19407-19434. https://doi.org/10.3390/molecules191219407
Bavishi K, Hatzakis NS. Shedding Light on Protein Folding, Structural and Functional Dynamics by Single Molecule Studies. Molecules. 2014; 19(12):19407-19434. https://doi.org/10.3390/molecules191219407
Chicago/Turabian StyleBavishi, Krutika, and Nikos S. Hatzakis. 2014. "Shedding Light on Protein Folding, Structural and Functional Dynamics by Single Molecule Studies" Molecules 19, no. 12: 19407-19434. https://doi.org/10.3390/molecules191219407
APA StyleBavishi, K., & Hatzakis, N. S. (2014). Shedding Light on Protein Folding, Structural and Functional Dynamics by Single Molecule Studies. Molecules, 19(12), 19407-19434. https://doi.org/10.3390/molecules191219407