The Regulation of Anthocyanin Synthesis in the Wheat Pericarp
Abstract
:1. Introduction
2. Results
2.1. Identification and Chromosome Location of Wheat Myc-Like Sequences
| Name | Alternative Name | Description | Pp | Plb | Pc | Rc | References | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -A1 | -B1 | -D1 | -A1 | -B1 | -D1 | -A1 | -B1 | -D1 | -A1 | -B1 | -D1 | ||||
| ► i:S29 Pp-A1pp-D1pp3 | “Saratovskaya 29” (“S29”) | Russian spring wheat | D | R | R | D | R | R | D | R | R | D | R | R | [25,33] |
| ► i:S29 Pp-A1Pp-D1Pp3PF* | i:S29 Pp1Pp2PF | wheat NIL developed on “S29”, donor—“Purple Feed” | D | R | D | D | R | D | D | R | D | D | R | D | [21,24,25] |
| ► i:S29 Pp-A1Pp-D1Pp3P* | i:S29 Pp1Pp3P | wheat NIL developed on “S29”, donor—“Purple” | D | R | D | D | R | D | D | R | D | D | R | D | [21,24,25] |
| ► i:S29 Pp-A1pp-D1Pp3PF** | no | wheat NIL developed on “S29”, donor—“Purple Feed” | D | R | R | D | R | R | D | R | R | D | R | R | [25] |
| ► i:S29 Pp-A1Pp-D1pp3PF | no | wheat NIL developed on “S29”, donor—“Purple Feed” | D | R | D | D | R | D | D | R | D | D | R | D | [25] |
| ► i:S29 Pp-A1pp-D1Pp3P** | no | wheat NIL developed on “S29”, donor—“Purple” | D | R | R | D | R | R | D | R | R | D | R | R | [25] |
| ► i:S29 Pp-A1Pp-D1pp3P | no | wheat NIL developed on “S29”, donor—“Purple” | D | R | D | D | R | D | D | R | D | D | R | D | [25] |
| ◄ i:S29 pp-A1pp-D1pp3 | line 140;“S29” (“YP” 4D*7A) | wheat NIL developed on “S29”, donor—“Yanetzkis Probat” | R | R | R | R | R | R | R | R | R | R | R | R | [25,33] |
| “Novosibirskaya 67” (“N67”) | no | Russian spring wheat | R | R | D | R | R | D | R | R | D | R | R | D | [24,34] |
| “Purple”* | no | Australian spring wheat “k-46990” | R | R | D | R | R | D | R | R | D | R | R | D | [24] |
| “Purple Feed”* | no | Canadian spring wheat “k-49426” | R | R | D | R | R | D | R | R | D | R | R | D | [24] |


2.2. Functional Activity of the Wheat Myc Gene Copies


2.3. The Full-Length Sequence of TaMyc1

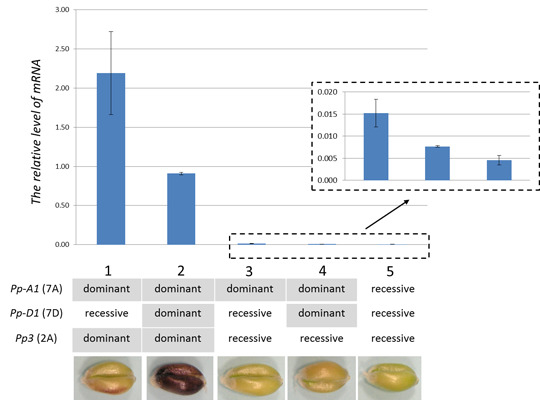
2.4. TaMyc1 Transcription as Affected by the Combination of Pp Alleles Present

3. Discussion
4. Experimental Section
4.1. Plant Materials
4.2. Gene Identification, Isolation and Sequence Analysis
4.3. Chromosomal Assignment of Wheat Myc Sequences
4.4. Transcription Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chalker-Scott, L. Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar]
- Hatier, J.H.B.; Gould, K.S. Anthocyanin function in vegetative organs. In Anthocyanins: Biosynthesis, Functions, and Applications; Gould, K., Davies, K., Winefield, C., Eds.; Springer Science+Business Media: New York, NY, USA, 2009; pp. 1–19. [Google Scholar]
- Wang, H.; Cao, G.; Prior, R.L. Oxygen radical absorbing capacity of anthocyanins. J. Agric. Food Chem. 1997, 45, 304–309. [Google Scholar]
- Hale, K.L.; McGrath, S.P.; Lombi, E.; Stack, S.M.; Terry, N.; Pickering, I.J.; George, G.N.; Pilon-Smits, E.A. Molybdenum sequestration in Brassica species. A role for anthocyanins? Plant Physiol. 2001, 126, 1391–1402. [Google Scholar]
- Hale, K.L.; Tufan, H.A.; Pickering, I.J.; George, G.N.; Terry, N.; Pilon, M.; Pilon-Smits, E.A.H. Anthocyanins facilitate tungsten accumulation in Brassica. Physiol. Plant. 2002, 116, 351–358. [Google Scholar]
- Manetas, Y. Why some leaves are anthocyanic and why most anthocyanic leaves are red? Flora 2006, 201, 163–177. [Google Scholar]
- Khlestkina, E.K. The adaptive role of flavonoids: Emphasis on cereals. Cereal Res. Commun. 2013, 41, 185–198. [Google Scholar]
- Dell’Agli, M.; Busciala, A.; Bosisio, E. Vascular effects of wine polyphenols. Cardiovasc. Res. 2004, 63, 593–602. [Google Scholar]
- Brown, J.E.; Kelly, M.F. Inhibition of lipid peroxidation by anthocyanins, anthocyanidins and their phenolic degradation products. Eur. J. Lipid Sci. Technol. 2007, 109, 66–71. [Google Scholar]
- Wang, H.; Nair, M.G.; Strasburg, G.M.; Chang, Y.C.; Booren, A.M.; Gray, J.I.; DeWitt, D.L. Antioxidant and antiinflammatory activities of anthocyanins and their aglycon, cyanidin, from tart cherries. J. Nat. Prod. 1999, 62, 294–296. [Google Scholar]
- Hui, C.; Bin, Y.; Xiaoping, Y.; Long, Y.; Chunye, C.; Mantian, M.; Wenhua, L. Anticancer activities of an anthocyanin-rich extract from black rice against breast cancer cells in vitro and in vivo. Nutr. Cancer 2010, 62, 1128–1136. [Google Scholar]
- Tsuda, T.; Horio, F.; Uchida, K.; Aoki, H.; Osawa, T. Dietary cyanidin 3-O-β-d-glucoside-rich purple corn color prevents obesity and ameliorates hyperglycemia. J. Nutr. 2003, 133, 2125–2130. [Google Scholar]
- Howard, B.V.; Kritchevsky, D. Phytochemicals and cardiovascular disease: A statement for healthcare professionals from the American heart association. Circulation 1997, 95, 2591–2593. [Google Scholar]
- Abdel-Aal, E.S.M.; Young, J.C.; Rabalski, I. Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J. Agric. Food Chem. 2006, 54, 4696–4704. [Google Scholar]
- Dykes, L.; Rooney, L.W. Phenolic compounds in cereal grains and their health benefits. Cereal Foods World 2006, 105–111. [Google Scholar]
- Zofajova, A.; Psenakova, I.; Havrlentova, M.; Piliarova, M. Accumulation of total anthocyanins in wheat grain. Agriculture (Poľnohospodárstvo) 2012, 58, 50–56. [Google Scholar]
- Ficco, D.B.M.; de simone, V.; Nigro, V.F.; Finocchiaro, F.; Papa, R.; de vita, P. Genetic variability in anthocyanin composition and nutritional properties of blue, purple and red bread (Triticum aestivum L.) and durum (Triticum turgidum L. spp. turgidum var. durum) wheats. J. Agric. Food Chem. 2014. [Google Scholar] [CrossRef]
- Li, W; Pickard, M.D.; Trust, B. Effect of thermal processing on antioxidant properties of purple wheat bran. Food Chem. 2008, 104, 1080–1086. [Google Scholar]
- Hirawan, R.; Diehl-Jones, W.; Trust, B. Comparative evaluation of the antioxidant potential of infant cereals produced from purple wheat and red rice grains and LC-MS analysis of their anthocyanins. J. Agric. Food Chem. 2011, 59, 12330–12341. [Google Scholar]
- Zeven, A.C. Wheats with purple and blue grains: A review. Euphytica 1991, 56, 243–258. [Google Scholar]
- Arbuzova, V.S.; Maystrenko, O.I.; Popova, O.M. Development of near-isogenic lines of the common wheat cultivar “Saratovskaya 29”. Cereal Res. Commun. 1998, 26, 39–46. [Google Scholar]
- Dobrovolskaya, O.B.; Arbuzova, V.S.; Lohwasser, U.; Röder, M.S.; Börner, A. Microsatellite mapping of complementary genes for purple grain colour in bread wheat (Triticum aestivum L.). Euphytica 2006, 150, 355–364. [Google Scholar]
- Khlestkina, E.K.; Röder, M.S.; Börner, A. Mapping genes controlling anthocyanin pigmentation on the glume and pericarp in tetraploid wheat (Triticum durum L.). Euphytica 2010, 171, 65–69. [Google Scholar]
- Tereshchenko, O.Y.; Gordeeva, E.I.; Arbuzova, V.S.; Börner, A.; Khlestkina, E.K. The D genome carries a gene determining purple grain colour in wheat. Cereal Res. Commun. 2012, 40, 334–341. [Google Scholar]
- Gordeeva, E.I.; Shoeva, O.Y.; Khlestkina, E.K. Marker-assisted development of bread wheat near-isogenic lines carrying various combinations of Pp (purple pericarp) alleles. Euphytica, 2015; in press. [Google Scholar]
- Saitoh, K.; Onishi, K.; Mikami, I.; Thidar, K.; Sano, Y. Allelic diversification at the C1 (OsC1) locus of wild and cultivated rice: Nucleotide changes associated with phenotypes. Genetics 2004, 168, 997–1007. [Google Scholar]
- Khlestkina, E.K. Genes determining coloration of different organs in wheat. Russ. J. Genet. Appl. Res. 2013, 3, 54–65. [Google Scholar]
- Li, W.L.; Faris, J.D.; Chittoor, J.M.; Leach, J.E.; Hulbert, S.H.; Liu, D.J.; Chen, P.D.; Gill, B.S. Genomic mapping of defense response genes in wheat. Theor. Appl. Genet. 1999, 98, 226–233. [Google Scholar]
- Hu, J.; Anderson, B.; Wessler, R. Isolation and characterization of rice R genes: Evidence for distinct evolutionary paths in rice and maize. Genetics 1996, 142, 1021–1031. [Google Scholar]
- Wang, C.; Shu, Q. Fine mapping and candidate gene analysis of purple pericarp gene Pb in rice (Oryza sativa L.). Chin. Sci. Bull. 2007, 52, 3097–3104. [Google Scholar]
- Ludwig, S.R.; Habera, L.F.; Dellaporta, S.L.; Wessler, S.R. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcription activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 1989, 86, 7092–7096. [Google Scholar]
- Tereshchenko, O.Y.; Arbuzova, V.S.; Khlestkina, E.K. Allelic state of the genes conferring purple pigmentation in different wheat organs predetermines transcriptional activity of the anthocyanin biosynthesis structural genes. J. Cereal Sci. 2013, 57, 10–13. [Google Scholar]
- Khlestkina, E.K.; Röder, M.S.; Pshenichnikova, T.A.; Börner, A. Functional diversity at the Rc (red coleoptile) gene in bread wheat. Mol. Breed. 2010, 25, 125–132. [Google Scholar]
- Khlestkina, E.K.; Pshenichnikova, T.A.; Röder, M.S.; Börner, A. Clustering anthocyanin pigmentation genes in wheat group 7 chromosomes. Cereal Res. Commun. 2009, 37, 391–398. [Google Scholar]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic Helix-Loop-Helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar]
- Goff, S.A.; Cone, K.C.; Chandler, V.L. Functional analysis of the transcriptional activator encoded by the maize B gene: Evidence for a direct functional of regulatory proteins. Genes Dev. 1992, 6, 864–875. [Google Scholar]
- Quattrocchio, F.; Wing, J.F.; van der Woude, K.; Mol, J.N.M.; Koes, R. Analysis of bHLH and MYB domain proteins: Species specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998, 13, 475–488. [Google Scholar]
- Cockram, J.; White, J.; Zuluaga, D.L.; Smith, D.; Comadran, J.; Macaulay, M.; Luo, Z.; Kearsey, M.J.; Werner, P.; Harrap, D.; et al. Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc. Natl. Acad. Sci. USA 2010, 107, 21611–21616. [Google Scholar]
- Chandler, V.L.; Radicella, J.P.; Robbins, T.P.; Chen, J.; Turks, D. Two regulatory genes of the maize anthocyanin pathway are homologous: Isolation of B utilizing R genomic sequences. Plant Cell 1989, 1, 1175–1183. [Google Scholar]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar]
- Tereshchenko, O.Y.; Pshenichnikova, T.A.; Salina, E.A.; Khlestkina, E.K. Development and molecular characterization of a novel wheat genotype having purple grain colour. Cereal Res. Commun. 2012, 40, 210–214. [Google Scholar]
- Mol, J.; Grotewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006, 46, 768–779. [Google Scholar]
- Dubos, C.; le Gourrierec, J.; Baudry, A.; Lanet, E.; Debeaujon, I.; Routaboul, J.-M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar]
- Schellmann, S.; Schnittger, A.; Kirik, V.; Wada, T.; Okada, K.; Beermann, A.; Thumfahrt, J.; Jürgens, G.; Hülskamp, M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002, 21, 5036–5046. [Google Scholar]
- Zhu, H.F.; Fitzsimmons, K.; Khandelwal, A.; Kranz, R.G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2009, 2, 790–802. [Google Scholar]
- Sears, E.R. Nullisomic analysis in common wheat. Am. Nat. 1953, 87, 245–252. [Google Scholar]
- Sears, E.R. Isochromosomes and telocentrics in Triticum vulgare. Genetics 1946, 31, 229–230. [Google Scholar]
- Endo, T.R.; Gill, B.S. The deletion stocks of common wheat. J. Hered. 1996, 87, 295–307. [Google Scholar]
- Plaschke, J.; Ganal, M.W.; Roder, M.S. Detection of genetic diversity in closely related bread wheat using microsatellite markers. Theor. Appl. Genet. 1995, 91, 1001–1007. [Google Scholar]
- Offerman, J.D.; Rychlik, W. Oligo primer analysis software. In Introduction to Bioinformatics: A Theoretical and Practical Approach; Krawetz, S.A., Womble, D.D., Eds.; Humana Press: New York, NY, USA, 2003; pp. 345–361. [Google Scholar]
- Wilkinson, P.A.; Winfield, M.O.; Barker, G.L.A.; Allen, A.M.; Burridge, A.; Coghill, J.A.; Burridge, A.; Edwards, K.J. CerealsDB 2.0: An integrated resource for plant breeders and scientists. BMC Bioinform. 2012, 13, 219. [Google Scholar]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucl. Acids Res. 1988, 16, 10881–10890. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7 (Suppl. 1), 10:1–10:12. [Google Scholar]
- Himi, E.; Nisar, A.; Noda, K. Colour genes (R and Rc) for grain and coleoptile upregulate flavonoid biosynthesis genes in wheat. Genome 2005, 48, 747–754. [Google Scholar]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar]
- Sample Availability: Samples of DNA of near-isogenic lines are available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shoeva, O.Y.; Gordeeva, E.I.; Khlestkina, E.K. The Regulation of Anthocyanin Synthesis in the Wheat Pericarp. Molecules 2014, 19, 20266-20279. https://doi.org/10.3390/molecules191220266
Shoeva OY, Gordeeva EI, Khlestkina EK. The Regulation of Anthocyanin Synthesis in the Wheat Pericarp. Molecules. 2014; 19(12):20266-20279. https://doi.org/10.3390/molecules191220266
Chicago/Turabian StyleShoeva, Olesya Y., Elena I. Gordeeva, and Elena K. Khlestkina. 2014. "The Regulation of Anthocyanin Synthesis in the Wheat Pericarp" Molecules 19, no. 12: 20266-20279. https://doi.org/10.3390/molecules191220266
APA StyleShoeva, O. Y., Gordeeva, E. I., & Khlestkina, E. K. (2014). The Regulation of Anthocyanin Synthesis in the Wheat Pericarp. Molecules, 19(12), 20266-20279. https://doi.org/10.3390/molecules191220266