Longevity Extension by Phytochemicals
Abstract
:1. Introduction
2. Phytochemicals Extend Lifespan in Evolutionarily Distant Heterotrophic Organisms by Targeting an Evolutionarily Conserved Set of Longevity-Defining Cellular Processes
2.1. Longevity-Extending Phytochemicals and Heterotrophic Organisms Whose Lifespans They Prolong
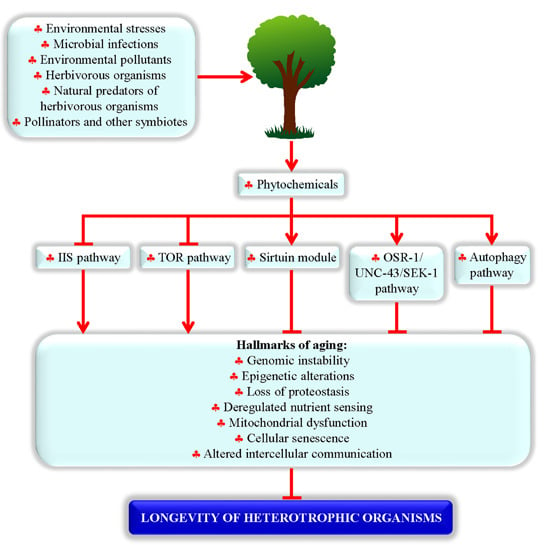
2.2. Proteins and Signaling Pathways Required for Longevity Extension by Phytochemicals
2.3. Processes Targeted by Longevity-Extending Phytochemicals in Evolutionarily Distant Organisms
| Phytochemical | Plant | Chemical Nature | Organism Exhibiting Lifespan Extension | Cellular Proteins and Signaling Pathways Required | Changes Caused |
|---|---|---|---|---|---|
| Acteoside | Phlomis anisodonta, Phlomis bruguieri, Verbascum phlomoides, Verbascum mallophorum, Buddleja globose, Buddleja cordata | Caffeoyl phenylethanoid glycoside (a phenolic compound) | The fruit fly Drosophila melanogaster [61] | NT | NT |
| Allicin | Allium sativum (garlic) | Organosulfur compound | • Senescence-accelerated mice [62,63,64,65] | NT | • Improved memory retention and acquisition [62,63,64,65] |
| Butein | Toxicodendron vernicifluum | Chalconoid (a phenolic compound) | • The yeast Saccharomyces cerevisiae [66] | • The sirtuin Sir1 [66] | NT |
| Caffeic acid, rosmarinic acid | Eucalyptus globulus, Salvinia molesta | Hydroxycinnamic acids (phenolic compounds) | • The nematode Caenorhabditis elegans [67] | • The OSR-1/UNC-43 (CaMKII)/SEK-1 (p38 MAPK) signaling pathway [67] • The sirtuin SIR-2.1 [67] • Caffeic acid only: the DAF-16/FOXO transcription factor [67] | • Lowered susceptibility to thermal stress [67] • Decreased oxidative damage to macromolecules [67] • Reduced body size, altered lipid metabolism, delayed reproductive timing [67] |
| Caffeine | Coffea plants | Methylxanthine (a purine) | • The yeasts Saccharomyces cerevisiae [68] and Schizosaccharomyces pombe [69] • The nematode C. elegans [70,71] | • In S. cerevisiae and Sch. pombe: the target of rapamycin complex 1 (TORC1) [68,69] • In C. elegans: the insulin-like receptor DAF-2, transcription factor DAF-16/FOXO and transcriptional activator CBP-1 [70,71] | • In S. cerevisiae: enhanced transcription of genes encoding heat-shock proteins and molecular chaperones [68] • In Sch. pombe: decelerated growth, G2 cell-cycle arrest, altered transcription of many nuclear genes, attenuated protein synthesis and inhibited phosphorylation of ribosomal S6 proteins [69] • In C. elegans: delayed onset of paralysis and reduced protein aggregation in nematode models of the Alzheimer’s and Huntington’s diseases [70,71] |
| Catechin | Vascular plants | Flavan-3-ol (a phenolic compound) | • The nematode C. elegans [72] | • The AKT-2 serine/threonine protein kinase, MEV-1 subunit of succinate-coenzyme Q oxidoreductase in the mitochondrial electron transport chain, and nuclear hormone receptor NHR-8 [72] | • Reduced body length and susceptibility to thermal stress [72] • Elevated pumping rate [72] |
| Celastrol | Tripterygium wilfordii, Celastrus regelii | Triterpenoid (a terpen) | • Transgenic mouse model of amyotrophic lateral sclerosis (ALS) [73] | NT | • Decelerated weight loss, improved motor performance, increased number of neurons and delayed onset of ALS [73] |
| Curcumin, tetrahydrocurcumin | Curcuma longa | Diarylheptanoids (phenolic compounds) | • The nematode C. elegans [74] • The fruit fly D. melanogaster, including 5 different models of Alzheimer’s disease [75,76,77] | • In C. elegans: the OSR-1/UNC-43 (CaMKII)/SEK-1 (p38 MAPK) signaling pathway [74] • In C. elegans: the sirtuin SIR-2.1 [74] • In C. elegans: the phosphatidylinositol 3-kinase AGE-1, transcription factor SKN-1/Nrf and MAPK kinase MEK-1 [74] | • In C. elegans: Reduced ROS levels, macromolecular oxidative damage, susceptibility to oxidative and thermal stresses, body length, and pumping rate [74] • In D. melanogaster: Decreased macromolecular oxidative damage, lowered susceptibility to oxidative stress, improved locomotor performance [75,76,77] |
| Crocin | Crocus, Gardenia | Carotenoid (a terpen) | • Dalton’s lymphoma ascites bearing mice [78] | NT | • Increased hemoglobin and lymphocytes [78] • Decreased white blood cell count and neutrophils [78] |
| Cryptotanshinone | Salvia miltiorrhiza | Tanshion (a quinone) | • The yeast S. cerevisiae [17] | • Mitochondrial superoxide dismutase Sod2, as well as the nutrient-sensing protein kinases Tor1, Sch9 and Gcn2 [17] | • Lowered ROS levels [17] |
| Cyanidin | Vitis vinifera, Vitis labrusca, Vaccinium myrtillus, Vaccinium uliginosum, Vaccinium alaskaense, Vaccinium angustifolium | Anthocyanidin (a phenolic compound) | • WI-38 human diploid fibroblasts [79] | NT | • Reduced oxidative damage to lipids and susceptibility to oxidative stress [79] |
| Diallyl trisulfide | Allium sativum (garlic) | A polysulfide (an organosulfide compound) | • The nematode C. elegans [80] | • The nicotinic acetylcholine receptor EAT-2 and transcription factor SKN-1/Nrf [80] | • Altered expression of many nuclear genes involved in metabolism and stress response [80] |
| Ellagic acid | Quercus alba, Quercus robur, Myriophyllum spicatum | Phenolic acid (a phenolic compound) | • The nematode C. elegans [81] | • The nicotinic acetylcholine receptor EAT-2 [81] | • Delayed beginning of egg deposition and reduced oxidative damage to water-soluble metabolites [81] |
| Epigallocatechin gallate | Camellia sinensis | Flavan-3-ol (a phenolic compound) | • The nematode C. elegans [82,83] | NT | • Lowered ROS levels, reduced susceptibility to oxidative stress, decreased oxidative damage to lipids, attenuated expression of nuclear genes encoding HSP-16, induced nuclear import of the transcription factor DAF-16/FOXO, reduced formation of Aβ deposits [82,83] |
| Epicatechin | Seed of Theobroma cacao, juice of Prunus domestica, seed of Vicia faba, oil from the fruit of Euterpe oleracea | Flavan-3-ol (a phenolic compound) | • The fruit fly Drosophila melanogaster [84] • Obese diabetic mice [84] | NT | • In obese diabetic mice: reduced degeneration of aortic vessels, lowered fat deposition, decreased hydropic degeneration in the liver, reduced markers of systematic inflammation, lowered serum LDL cholesterol, decreased level of circulating insulin-like growth factor 1, improved skeletal muscle stress output, increased concentration of hepatic glutathione, elevated superoxide dismutase activity, amplified AMP-activated protein kinase activity in the liver and skeletal muscle [84] |
| Ferulsinaic acid | Ferula plants | Sesquiterpene coumarin (a terpene) | • The nematode C. elegans [85] | NT | • Reduced susceptibility to oxidative and thermal stresses, decreased oxidative damage to lipids, lowered formation of advanced glycation end products [85] |
| Fisetin | Acacia greggii, Acacia berlandieri, Butea frondosa, Gleditsia triacanthos, Quebracho colorado, Rhus cotinus | Flavonol (a phenolic compound) | • The yeast S. cerevisiae [66] • The nematode C. elegans [86] | • In S. cerevisiae: The sirtuin Sir1 [66] • In C. elegans: Nuclear import of the transcription factor DAF-16/FOXO [86] | • In S. cerevisiae: NT [66] • In C. elegans: Lowered ROS levels, reduced susceptibility to oxidative stress, decreased oxidative damage to macromolecules, induced nuclear import of transcription factor DAF-16/FOXO [86] |
| Gallic acid | Quercus alba, Quercus robur, Caesalpinia mimosoides, Boswellia dalzielii, Rhodiola rosea, Toona sinensis | Phenolic acid (a phenolic compound) | • The nematode C. elegans [81] | • The nicotinic acetylcholine receptor EAT-2 [81] | • Increased body length, delayed beginning of egg deposition and reduced oxidative damage to water-soluble metabolites [81] |
| Glaucarubinone | Simaroubaceae plants | Triterpenoid (a terpen) | • The nematode C. elegans [87] | NT | • Increased rate of oxygen consumption and lowered levels of neutral lipids [87] |
| HDTIC-1, HDTIC-2 | Astragalus membranceus | Indolizines (indole compounds) | • Human fetal lung diploid fibroblasts [88] | NT | • Improved growth and proliferation, accelerated entry from G0 or G1 phase to S phase, decreased activity of the senescence-associated-β-galactosidase, and reduced formation of advanced glycation end products [88] |
| Icariin, icariside II | Epimedium plants | Flavonol glycosides (phenolic compounds) | • The nematode C. elegans [89] | • The insulin-like receptor DAF-2, transcription factor DAF-16/FOXO and heat shock transcription factor HSF-1 [89] | • Reduced susceptibility to oxidative and thermal stresses, decelerated decline in age-related locomotion, delayed onset of paralysis caused by the proteotoxicity of polyQ and Aβ(1–42), enhanced transcription of the SOD-3 and HSP-12.3 genes [89] |
| Kaempferol | Aloe vera, Coccinia grandis, Cuscuta chinensis, Euphorbia pekinensis, Glycine max, Hypericum perforatum | Flavonol (a phenolic compound) | • The nematode C. elegans [86] | • Nuclear import of the transcription factor DAF-16/FOXO [86] | • Lowered ROS levels, reduced susceptibility to oxidative stress, decreased oxidative damage to macromolecules, induced nuclear import of transcription factor DAF-16/FOXO [86] |
| Myricetin | Morella rubra, Myrica cerifera, Rosa damascene, Salvia hispanica, Hovenia dulcis, Ceratonia siliqua | Flavonol (a phenolic compound) | • The nematode C. elegans [90] | • Nuclear import of the transcription factor DAF-16/FOXO [90] | • Lowered ROS levels, reduced oxidative damage to proteins, induced nuclear import of transcription factor DAF-16/FOXO, enhanced transcription of the SOD-3 gene [90] |
| Nordihydroguaiaretic acid | Larrea tridentata | Lignan (a phenolic compound) | • Transgenic mouse model of ALS [91] • Male mice [92] • Rats [93] • The fruit fly D. melanogaster [94] • Mosquitoes [95] | NT | • In transgenic mouse model of ALS: reduced motor dysfunction [91] • In D. melanogaster: lowered rate of oxygen consumption [94] |
| Oleuropein | Olea europaea | Phenylethanoid (a phenolic compound) | • Human embryonic fibroblasts [96] | NT | • Lowered ROS levels, reduced oxidative damage to proteins, increased rate of proteasomal degradation of oxidatively damaged proteins, decelerated age-related decline in proteasome activity [96] |
| Phloridzin | Pyrus communis, Malus domestica, Prunus avium, Rosaceae plants, Dianthus caryophyllus | Chalconoid (a phenolic compound) | • The yeast S. cerevisiae [97] | • Cytosolic and mitochondrial superoxide dismutases Sod1 and Sod2 (respectively) [97] | • Lowered ROS levels, decreased susceptibility to oxidative stress, activated transcription of the SOD1, SOD2 and SIR2 genes, increased superoxide dismutase activity [97] |
| Quercetin, Q3'G, Q3M, isorhamnetin, tamarixetin | Capparis spinosa, Levisticum officinale, Rumex acetosa, Raphanus sativus, Ceratonia siliqua, Anethum graveolens | Flavonols (phenolic compounds) | • The yeast S. cerevisiae [98] • The nematode C. elegans [67,99,100,101,103] • Human embryonic fibroblasts [102] | • In C. elegans: the insulin-like receptor DAF-2, phosphatidylinositol 3-kinase AGE-1 and nuclear import of the transcription factor DAF-16/FOXO [99,100,101], as well as the OSR-1/UNC-43 (CaMKII)/SEK-1 (p38 MAPK) signaling pathway [101] | • In
S. cerevisiae: Lowered ROS levels, decreased glutathione oxidation, reduced protein carbonylation, lowered lipid peroxidation, decreased susceptibility to oxidative stress [98] • In C. elegans: Lowered ROS levels, reduced oxidative damage to macromolecules, enhanced anti-oxidative activities, decreased susceptibility to thermal and oxidative stresses, lowered level of neutral lipids, induced nuclear import of transcription factor DAF-16/FOXO [67,99,100,101] • In human fibroblasts: Lowered activity of the senescence-associated-β-galactosidase, decreased ROS levels, reduced susceptibility to oxidative stress, increased proteasome activity [102] |
| Reserpine | Rauvolfia serpentina | Indole alkaloid (an indole compound) | • The nematode
C. elegans [104] • The nematode C. elegans model of Alzheimer’s disease [105] | • TPH-1, a tryptophan hydroxlase enzyme [104]. | • Reduced susceptibility to thermal stress, decelerated decline in age-related locomotion and pharyngeal pumping, delayed postembryonic development [104,105] • The nematode C. elegans model of Alzheimer’s disease: delayed onset of paralysis caused by the proteotoxicity of Aβ [105] |
| Resveratrol | Vitis plants, Vaccinium alaskaense, Vaccinium angustifolium, Rubus idaeus, Rubus occidentalis, Broussonetia papyrifera | Stilbenoid (a phenolic compound) | • The yeast
S. cerevisiae [66] • The nematode C. elegans [106] • The fruit fly D. melanogaster [106] • The short-lived fish Nothobranchius Furzeri [107] • The honey bee Apis mellifera [108] • Mice on a high-calorie diet [109] | • In
S. cerevisiae: the sirtuin Sir1 [66] • The nematode C. elegans: the sirtuin SIR-2.1 [106] • The fruit fly D. melanogaster: the sirtuin Sir2 [106] • Mouse: SIRT1 and many other cellular targets, direct or indirect [42,54,60] | • In
S. cerevisiae: reduced frequency of rDNA recombination [66] • In the nematode C. elegans: induced autophagy [110] • In N. furzeri: delayed age-related decay of locomotor activity and cognitive performances, reduced neurofibrillary degeneration in the brain [107] • In mouse: increased insulin sensitivity, increased activities of AMP-activated protein kinase (AMPK) and peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC-1α), reduced levels of insulin-like growth factor-1 (IGF-I), increased number of mitochondria, altered transcription of many nuclear genes [109] |
| Spermidine, putrescine | Wheat (Triticum spp.) germ, Ipomoea batatas, Pisum sativum, Glycine max, Glycine soja | Polyamines | • The yeast S. cerevisiae [111] • The nematode C. elegans [111] • The fruit fly D. melanogaster [111] • Human peripheral blood mononuclear cells (PBMC) [111] | • In S. cerevisiae, C. elegans and D. melanogaster: autophagy [111,112] | • In S. cerevisiae, D. melanogaster and PBMC: lowered activities of histone acetyltransferases, increased histone H3 deacetylation, activated transcription of many autophagy-related genes, induced autophagy, delayed onset of age-related necrotic cell death, reduced age-related decline of locomotor activity [111] • In D. melanogaster: decelerated age-related decline of locomotor activity, increased level of triglycerides, altered relative levels of fatty acid species and phospholipid classes [112,113] |
| Tannic acid | Caesalpinia spinosa, Rhus semialata, Quercus infectoria, Rhus coriaria | Polyphenol (a phenolic compound) | • The nematode C. elegans [70,81,114] | • The mitogen-activated protein kinase kinase SEK-1, transcription factor DAF-16/FOXO, nicotinic acetylcholine receptor EAT-2 and MEV-1 subunit of succinate-coenzyme Q oxidoreductase in the mitochondrial electron transport chain [70,81,114] | • Reduced body length, decreased susceptibility to thermal and oxidative stresses, lowered levels of triglycerides, enhanced anti-oxidant capacity [70,81,114] |
| Tyrosol | Oil from the fruit of Olea europaea, oil from the kernels of Argania spinosa, leaves from Camellia sinensis | Phenylethanoid (a phenolic compound) | • The nematode C. elegans [115] | • The insulin-like receptor DAF-2, transcription factor DAF-16/FOXO and heat shock transcription factor HSF-1 [115] | • Decreased susceptibility to thermal and oxidative stresses, decelerated onset of age-related decline in pharyngeal pumping, activated transcription of nuclear genes encoding several heat-shock proteins [115] |
2.3.1. Yeasts
2.3.2. The Nematode C. elegans
2.3.3. The Fruit Fly D. melanogaster
2.3.4. The Fish Nothobranchius Furzeri
2.3.5. Laboratory Mouse
2.3.6. Cultured Human Cells
2.4. Mechanisms of Longevity Extension by Phytochemicals Are Evolutionarily Conserved
3. Phytochemicals: Interspecies Chemical Signals That May Contribute to the Evolution of Longevity Regulation Mechanisms within Natural Ecosystems
3.1. The “Xenohormesis” Hypothesis
3.2. Many Observations Contradict the Xenohormesis Hypothesis
3.3. An Extended Hypothesis on the Role of Phytochemicals in the Ecosystemic Evolution of Longevity Regulation Mechanisms
4. Conclusions and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Harborne, J.R. Introduction to Ecological Biochemistry, 4th ed.; Elsevier Inc.: London, UK, 1993; p. 316. [Google Scholar]
- Gershenzon, J. The cost of plant chemical defense against herbivory: A biochemical perspective. In Insect-Plant Interactions; Bernays, E.A., Ed.; CRC Press: Boca Raton, USA, 1994; pp. 105–173. [Google Scholar]
- Reymond, P.; Weber, H.; Damond, M.; Farmer, E.E. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 2000, 12, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Hermsmeier, D.; Schittko, U.; Baldwin, I.T. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. I. Large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol. 2001, 125, 683–700. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.O.; Wightman, E.L. Herbal extracts and phytochemicals: Plant secondary metabolites and the enhancement of human brain function. Adv. Nutr. 2011, 2, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Strobel, G.; Daisy, B.; Castillo, U.; Harper, J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004, 67, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.C.; Kharwar, R.N.; Strobel, G.A. Chemical and functional diversity of natural products from plant associated endophytic fungi. Nat. Prod. Commun. 2009, 4, 1511–1532. [Google Scholar] [PubMed]
- Yu, H.; Zhang, L.; Li, L.; Zheng, C.; Guo, L.; Li, W.; Sun, P.; Qin, L. Recent developments and future prospects of antimicrobial metabolites produced by endophytes. Microbiol. Res. 2010, 165, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Bascom-Slack, C.A.; Arnold, A.E.; Strobel, S.A. IBI series winner. Student-directed discovery of the plant microbiome and its products. Science 2012, 338, 485–486. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.H.; Debbab, A.; Proksch, P. Fungal endophytes—Secret producers of bioactive plant metabolites. Pharmazie 2013, 68, 499–505. [Google Scholar] [PubMed]
- Mousa, W.K.; Raizada, M.N. The diversity of anti-microbial secondary metabolites produced by fungal endophytes: An interdisciplinary perspective. Front. Microbiol. 2013, 4, 65. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.G.; Halkier, B.A. New insight into the biosynthesis and regulation of indole compounds in Arabidopsis thaliana. Planta 2005, 221, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Hooper, P.L.; Hooper, P.L.; Tytell, M.; Vígh, L. Xenohormesis: Health benefits from an eon of plant stress response evolution. Cell Stress Chaperones 2010, 15, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Higdon, J.; Drake, V.J. An Evidence-Based Approach to Phytochemicals and Other Dietary Factors, 2nd ed.; Thieme: New York, NY, USA, 2012; p. 328. [Google Scholar]
- Menendez, J.A.; Joven, J.; Aragonès, G.; Barrajón-Catalán, E.; Beltrán-Debón, R.; Borrás-Linares, I.; Camps, J.; Corominas-Faja, B.; Cufí, S.; Fernández-Arroyo, S.; et al. Xenohormetic and anti-aging activity of secoiridoid polyphenols present in extra virgin olive oil: A new family of gerosuppressant agents. Cell Cycle 2013, 12, 555–578. [Google Scholar] [CrossRef] [PubMed]
- Si, H.; Liu, D. Dietary antiaging phytochemicals and mechanisms associated with prolonged survival. J. Nutr. Biochem. 2014, 25, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Song, L.; Liu, S.Q.; Huang, D. Tanshinones extend chronological lifespan in budding yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2014, 98, 8617–8628. [Google Scholar] [CrossRef] [PubMed]
- Somani, S.J.; Modi, K.P.; Majumdar, A.S.; Sadarani, B.N. Phytochemicals and their potential usefulness in inflammatory bowel disease. Phytother. Res. 2015, 29, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 2003, 64, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Tahara, S. A journey of twenty-five years through the ecological biochemistry of flavonoids. Biosci. Biotechnol. Biochem. 2007, 71, 1387–1404. [Google Scholar] [CrossRef] [PubMed]
- Murakami, A. Modulation of protein quality control systems by food phytochemicals. J. Clin. Biochem. Nutr. 2013, 52, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Adrian, M.; Jeandet, P.; Veneau, J.; Weston, L.A.; Bessis, R. Biological activity of resveratrol, a stilbenic compound from grapevines, against Botrytis cinerea, the causal agent for gray mold. J. Chem. Ecol. 1997, 23, 1689–1702. [Google Scholar] [CrossRef]
- Heath, M.C. Hypersensitive response-related death. Plant Mol. Biol. 2000, 44, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Trewavas, A.; Stewart, D. Paradoxical effects of chemicals in the diet on health. Curr. Opin. Plant Biol. 2003, 6, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Arimura, G.; Kost, C.; Boland, W. Herbivore-induced, indirect plant defences. Biochim. Biophys. Acta 2005, 1734, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Brencic, A.; Winans, S.C. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 2005, 69, 155–194. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P.; Son, T.G.; Camandola, S. Viewpoint: Mechanisms of action and therapeutic potential of neurohormetic phytochemicals. Dose Response 2007, 5, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Santiago, R.; Malvar, R.A. Role of dehydrodiferulates in maize resistance to pests and diseases. Int. J. Mol. Sci. 2010, 11, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhan, J.C.; Yang, H.R.; Huang, W.D. Changes of resveratrol and antioxidant enzymes during UV-induced plant defense response in peanut seedlings. J. Plant Physiol. 2010, 167, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Arimura, G.; Ozawa, R.; Maffei, M.E. Recent advances in plant early signaling in response to herbivory. Int. J. Mol. Sci. 2011, 12, 3723–3739. [Google Scholar] [CrossRef] [PubMed]
- Barros-Rios, J.; Malvar, R.A.; Jung, H.J.; Santiago, R. Cell wall composition as a maize defense mechanism against corn borers. Phytochemistry 2011, 72, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P. Sulfur-containing secondary metabolites from Arabidopsis thaliana and other Brassicaceae with function in plant immunity. ChemBioChem 2012, 13, 1846–1859. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Slusarenko, A.J.; Gruhlke, M.C. Sulfur and sulfur compounds in plant defence. Nat. Prod. Commun. 2012, 7, 395–400. [Google Scholar] [PubMed]
- Huot, O.B.; Nachappa, P.; Tamborindeguy, C. The evolutionary strategies of plant defenses have a dynamic impact on the adaptations and interactions of vectors and pathogens. Insect Sci. 2013, 20, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Lyons, R. Intervention of phytohormone pathways by pathogen effectors. Plant Cell 2014, 26, 2285–2309. [Google Scholar] [CrossRef] [PubMed]
- Porras-Alfaro, A.; Bayman, P. Hidden fungi, emergent properties: Endophytes and microbiomes. Annu. Rev. Phytopathol. 2011, 49, 291–315. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shan, T.; Mou, Y.; Zhou, L. Plant-derived bioactive compounds produced by endophytic fungi. Mini Rev. Med. Chem. 2011, 11, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Kusari, S.; Hertweck, C.; Spiteller, M. Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 2012, 19, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.; Raghunatha, P.; Joshi, S.R. Diversity and biological activities of endophytic fungi of Emblica officinalis, an ethnomedicinal plant of India. Mycobiology 2012, 40, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, T.; Ming, Q.; Wu, L.; Rahman, K.; Qin, L. Alkaloids produced by endophytic fungi: A review. Nat. Prod. Commun. 2012, 7, 963–968. [Google Scholar] [PubMed]
- Lebeis, S.L. The potential for give and take in plant-microbiome relationships. Front. Plant Sci. 2014, 5, 287. [Google Scholar] [CrossRef] [PubMed]
- Harikumar, K.B.; Aggarwal, B.B. Resveratrol: A multitargeted agent for age-associated chronic diseases. Cell Cycle 2008, 7, 1020–1035. [Google Scholar] [CrossRef] [PubMed]
- Son, T.G.; Camandola, S.; Mattson, M.P. Hormetic dietary phytochemicals. Neuromol. Med. 2008, 10, 236–246. [Google Scholar] [CrossRef]
- Calabrese, V.; Cornelius, C.; Dinkova-Kostova, A.T.; Iavicoli, I.; Di Paola, R.; Koverech, A.; Cuzzocrea, S.; Rizzarelli, E.; Calabrese, E.J. Cellular stress responses, hormetic phytochemicals and vitagenes in aging and longevity. Biochim. Biophys. Acta 2012, 1822, 753–783. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Guha, S.; Sun, X.; Cao, M.; Wang, X.; Zou, S. Nutraceutical interventions for promoting healthy aging in invertebrate models. Oxid. Med. Cell. Longev. 2012, 2012, 718491. [Google Scholar] [CrossRef] [PubMed]
- Lamming, D.W.; Sabatini, D.M.; Baur, J.A. Pharmacologic means of extending lifespan. J. Clin. Exp. Pathol. 2012, 4, 7327. [Google Scholar] [PubMed]
- Vauzour, D. Dietary polyphenols as modulators of brain functions: Biological actions and molecular mechanisms underpinning their beneficial effects. Oxid. Med. Cell. Longev. 2012, 2012, 914273. [Google Scholar] [CrossRef] [PubMed]
- Argyropoulou, A.; Aligiannis, N.; Trougakos, I.P.; Skaltsounis, A.L. Natural compounds with anti-ageing activity. Nat. Prod. Rep. 2013, 30, 1412–1437. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Khor, T.O.; Shu, L.; Su, Z.Y.; Fuentes, F.; Kong, A.N. Dietary phytochemicals and cancer prevention: Nrf2 signaling, epigenetics, and cell death mechanisms in blocking cancer initiation and progression. Pharmacol. Ther. 2013, 137, 153–171. [Google Scholar] [CrossRef] [PubMed]
- Lucanic, M.; Lithgow, G.J.; Alavez, S. Pharmacological lifespan extension of invertebrates. Ageing Res. Rev. 2013, 12, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Monroy, A.; Lithgow, G.J.; Alavez, S. Curcumin and neurodegenerative diseases. Biofactors 2013, 39, 122–132. [Google Scholar] [CrossRef] [PubMed]
- De Cabo, R.; Carmona-Gutierrez, D.; Bernier, M.; Hall, M.N.; Madeo, F. The search for antiaging interventions: From elixirs to fasting regimens. Cell 2014, 157, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Grabacka, M.M.; Gawin, M.; Pierzchalska, M. Phytochemical modulators of mitochondria: The search for chemopreventive agents and supportive therapeutics. Pharmaceuticals 2014, 7, 913–942. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, B.P.; Sinclair, D.A. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol. Sci. 2014, 35, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.O. Polyphenols and the human brain: Plant “secondary metabolite” ecologic roles and endogenous signaling functions drive benefits. Adv. Nutr. 2014, 5, 515–533. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Havermann, S.; Büchter, C.; Wätjen, W. Caenorhabditis elegans as model system in pharmacology and toxicology: Effects of flavonoids on redox-sensitive signalling pathways and ageing. ScientificWorldJournal 2014, 2014, 920398. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jo, D.G.; Park, D.; Chung, H.Y.; Mattson, M.P. Adaptive cellular stress pathways as therapeutic targets of dietary phytochemicals: Focus on the nervous system. Pharmacol. Rev. 2014, 66, 815–868. [Google Scholar] [CrossRef] [PubMed]
- Mansuri, M.L.; Parihar, P.; Solanki, I.; Parihar, M.S. Flavonoids in modulation of cell survival signalling pathways. Genes Nutr. 2014, 9, 400. [Google Scholar] [CrossRef] [PubMed]
- Rege, S.D.; Geetha, T.; Griffin, G.D.; Broderick, T.L.; Babu, J.R. Neuroprotective effects of resveratrol in Alzheimer disease pathology. Front. Aging Neurosci. 2014, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.A.; Guarente, L. Small-molecule allosteric activators of sirtuins. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 363–380. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Jiang, S.; Luo, P.; Wu, J.; Gao, P. Isolation, purification and structure identification of antioxidant compound from the roots of Incarvillea younghusbandii Sprague and its life span prolonging effect in Drosophila melanogaster. Nat. Prod. Res. 2008, 22, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, T.; Takashina, K.; Chu, P.J.; Saito, H.; Nishiyama, N. Prolongation of life span and improved learning in the senescence accelerated mouse produced by aged garlic extract. Biol. Pharm. Bull. 1994, 17, 1589–1594. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, T.; Saito, H.; Nishiyama, N. Aged garlic extract prolongs longevity and improves spatial memory deficit in senescence-accelerated mouse. Biol. Pharm. Bull. 1996, 19, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, T.; Saito, H.; Nishiyama, N. Anti-ageing effect of aged garlic extract in the inbred brain atrophy mouse model. Clin. Exp. Pharmacol. Physiol. 1997, 24, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, N.; Moriguchi, T.; Saito, H. Beneficial effects of aged garlic extract on learning and memory impairment in the senescence-accelerated mouse. Exp. Gerontol. 1997, 32, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; Scherer, B.; Sinclair, D.A. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, K.; Saul, N.; Chakrabarti, S.; Stürzenbaum, S.R.; Menzel, R.; Steinberg, C.E. Hormetins, antioxidants and prooxidants: Defining quercetin-, caffeic acid- and rosmarinic acid-mediated life extension in C. elegans. Biogerontology 2011, 12, 329–347. [Google Scholar] [CrossRef] [PubMed]
- Wanke, V.; Cameroni, E.; Uotila, A.; Piccolis, M.; Urban, J.; Loewith, R.; De Virgilio, C. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 2008, 69, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Rallis, C.; Codlin, S.; Bähler, J. TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell 2013, 12, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Lublin, A.; Isoda, F.; Patel, H.; Yen, K.; Nguyen, L.; Hajje, D.; Schwartz, M.; Mobbs, C. FDA-approved drugs that protect mammalian neurons from glucose toxicity slow aging dependent on Cbp and protect against proteotoxicity. PLoS ONE 2011, 6, e27762. [Google Scholar] [CrossRef] [PubMed]
- Sutphin, G.L.; Bishop, E.; Yanos, M.E.; Moller, R.M.; Kaeberlein, M. Caffeine extends life span, improves healthspan, and delays age-associated pathology in Caenorhabditis elegans. Longev. Healthspan 2012, 1, 9. [Google Scholar] [CrossRef] [PubMed]
- Saul, N.; Pietsch, K.; Menzel, R.; Stürzenbaum, S.R.; Steinberg, C.E. Catechin induced longevity in C. elegans: From key regulator genes to disposable soma. Mech. Ageing Dev. 2009, 130, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Kiaei, M.; Kipiani, K.; Petri, S.; Chen, J.; Calingasan, N.Y.; Beal, M.F. Celastrol blocks neuronal cell death and extends life in transgenic mouse model of amyotrophic lateral sclerosis. Neurodegener. Dis. 2005, 2, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Liao, V.H.; Yu, C.W.; Chu, Y.J.; Li, W.H.; Hsieh, Y.C.; Wang, T.T. Curcumin-mediated lifespan extension in Caenorhabditis elegans. Mech. Ageing Dev. 2011, 132, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Lee, B.S.; Semnani, S.; Avanesian, A.; Um, C.Y.; Jeon, H.J.; Seong, K.M.; Yu, K.; Min, K.J.; Jafari, M. Curcumin extends life span, improves health span, and modulates the expression of age-associated aging genes in Drosophila melanogaster. Rejuvenation Res. 2010, 13, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.R.; Xiao, F.; Yuan, P.; Chen, Y.; Gao, Q.K.; Parnell, L.D.; Meydani, M.; Ordovas, J.M.; Li, D.; Lai, C.Q. Curcumin-supplemented diets increase superoxide dismutase activity and mean lifespan in Drosophila. Age 2013, 35, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Caesar, I.; Jonson, M.; Nilsson, K.P.; Thor, S.; Hammarström, P. Curcumin promotes A-beta fibrillation and reduces neurotoxicity in transgenic Drosophila. PLoS ONE 2012, 7, e31424. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, H.A.; Sam, S.; Feroz, A.; Ravesh, Z.; Shah, G.A.; Sharma, M. Crocin from Kashmiri saffron (Crocus sativus) induces in vitro and in vivo xenograft growth inhibition of Dalton's lymphoma (DLA) in mice. Asian Pac. J. Cancer Prev. 2009, 10, 887–890. [Google Scholar] [PubMed]
- Choi, M.J.; Kim, B.K.; Park, K.Y.; Yokozawa, T.; Song, Y.O.; Cho, E.J. Anti-aging effects of cyanidin under a stress-induced premature senescence cellular system. Biol. Pharm. Bull. 2010, 33, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Powolny, A.A.; Singh, S.V.; Melov, S.; Hubbard, A.; Fisher, A.L. The garlic constituent diallyl trisulfide increases the lifespan of C. elegans via skn-1 activation. Exp. Gerontol. 2011, 46, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Saul, N.; Pietsch, K.; Stürzenbaum, S.R.; Menzel, R.; Steinberg, C.E. Diversity of polyphenol action in Caenorhabditis elegans: Between toxicity and longevity. J. Nat. Prod. 2011, 74, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.; Wink, M. Epigallocatechin gallate from green tea (Camellia sinensis) increases lifespan and stress resistance in Caenorhabditis elegans. Planta Med. 2009, 75, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.; Wink, M. Epigallocatechin gallate inhibits beta amyloid oligomerization in Caenorhabditis elegans and affects the daf-2/insulin-like signaling pathway. Phytomedicine 2010, 17, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Si, H.; Fu, Z.; Babu, P.V.; Zhen, W.; Leroith, T.; Meaney, M.P.; Voelker, K.A.; Jia, Z.; Grange, R.W.; Liu, D. Dietary epicatechin promotes survival of obese diabetic mice and Drosophila melanogaster. J. Nutr. 2011, 141, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.A. Ferulsinaic acid attenuation of advanced glycation end products extends the lifespan of Caenorhabditis elegans. J. Pharm. Pharmacol. 2011, 63, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Kampkötter, A.; Nkwonkam, C.G.; Zurawski, R.F.; Timpel, C.; Chovolou, Y.; Wätjen, W.; Kahl, R. Effects of the flavonoids kaempferol and fisetin on thermotolerance, oxidative stress and FoxO transcription factor DAF-16 in the model organism Caenorhabditis elegans. Arch. Toxicol. 2007, 81, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Zarse, K.; Bossecker, A.; Müller-Kuhrt, L.; Siems, K.; Hernandez, M.A.; Berendsohn, W.G.; Birringer, M.; Ristow, M. The phytochemical glaucarubinone promotes mitochondrial metabolism, reduces body fat, and extends lifespan of Caenorhabditis elegans. Horm. Metab. Res. 2011, 43, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, Z.; Ma, X.; Huang, Y.; Liu, X.; Tu, P.; Tong, T. HDTIC-1 and HDTIC-2, two compounds extracted from Astragali Radix, delay replicative senescence of human diploid fibroblasts. Mech. Ageing Dev. 2003, 124, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.J.; Huang, J.H.; Zhang, S.Q.; Wu, B.; Kapahi, P.; Zhang, X.M.; Shen, Z.Y. Icariin and its derivative icariside II extend healthspan via insulin/IGF-1 pathway in C. elegans. PLoS ONE 2011, 6, e28835. [Google Scholar] [CrossRef] [PubMed]
- Grünz, G.; Haas, K.; Soukup, S.; Klingenspor, M.; Kulling, S.E.; Daniel, H.; Spanier, B. Structural features and bioavailability of four flavonoids and their implications for lifespan-extending and antioxidant actions in C. elegans. Mech. Ageing Dev. 2012, 133, 1–10. [Google Scholar] [CrossRef] [PubMed]
- West, M.; Mhatre, M.; Ceballos, A.; Floyd, R.A.; Grammas, P.; Gabbita, S.P.; Hamdheydari, L.; Mai, T.; Mou, S.; Pye, Q.N.; et al. The arachidonic acid 5-lipoxygenase inhibitor nordihydroguaiaretic acid inhibits tumor necrosis factor alpha activation of microglia and extends survival of G93A-SOD1 transgenic mice. J. Neurochem. 2004, 91, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Strong, R.; Miller, R.A.; Astle, C.M.; Floyd, R.A.; Flurkey, K.; Hensley, K.L.; Javors, M.A.; Leeuwenburgh, C.; Nelson, J.F.; Ongini, E.; et al. Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell 2008, 7, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Buu-Hoi, N.P.; Ratsimamanga, A.R. Retarding action of nordihydroguaiaretic acid on aging in the rat. C. R. Seances Soc. Biol. Fil. 1959, 153, 1180–1182. [Google Scholar] [PubMed]
- Miquel, J.; Fleming, J.; Economos, A.C. Antioxidants, metabolic rate and aging in Drosophila. Arch. Gerontol. Geriatr. 1982, 1, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Richie, J.P.; Mills, B.J.; Lang, C.A. Dietary nordihydroguaiaretic acid increases the life span of the mosquito. Proc. Soc. Exp. Biol. Med. 1986, 183, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Katsiki, M.; Chondrogianni, N.; Chinou, I.; Rivett, A.J.; Gonos, E.S. The olive constituent oleuropein exhibits proteasome stimulatory properties in vitro and confers life span extension of human embryonic fibroblasts. Rejuvenation Res. 2007, 10, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Sun, K.; Lu, J.; Weng, Y.; Taoka, A.; Sakagami, Y.; Qi, J. Anti-aging effects of phloridzin, an apple polyphenol, on yeast via the SOD and Sir2 genes. Biosci. Biotechnol. Biochem. 2011, 75, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Belinha, I.; Amorim, M.A.; Rodrigues, P.; de Freitas, V.; Moradas-Ferreira, P.; Mateus, N.; Costa, V. Quercetin increases oxidative stress resistance and longevity in Saccharomyces cerevisiae. J. Agric. Food Chem. 2007, 55, 2446–2451. [Google Scholar] [CrossRef] [PubMed]
- Kampkötter, A.; Nkwonkam, C.G.; Zurawski, R.F.; Timpel, C.; Chovolou, Y.; Wätjen, W.; Kahl, R. Investigations of protective effects of the flavonoids quercetin and rutin on stress resistance in the model organism Caenorhabditis elegans. Toxicology 2007, 234, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Kampkötter, A.; Timpel, C.; Zurawski, R.F.; Ruhl, S.; Chovolou, Y.; Proksch, P.; Wätjen, W. Increase of stress resistance and lifespan of Caenorhabditis elegans by quercetin. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 149, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, K.; Saul, N.; Menzel, R.; Stürzenbaum, S.R.; Steinberg, C.E. Quercetin mediated lifespan extension in Caenorhabditis elegans is modulated by age-1, daf-2, sek-1 and unc-43. Biogerontology 2009, 10, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Chondrogianni, N.; Kapeta, S.; Chinou, I.; Vassilatou, K.; Papassideri, I.; Gonos, E.S. Anti-ageing and rejuvenating effects of quercetin. Exp. Gerontol. 2010, 45, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.L.; Ahiko, T.; Miyakawa, T.; Amino, H.; Hu, F.; Furihata, K.; Kita, K.; Shirasawa, T.; Sawano, Y.; Tanokura, M. Isolation and Caenorhabditis elegans lifespan assay of flavonoids from onion. J. Agric. Food Chem. 2011, 59, 5927–5934. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Arya, U.; SoundaraRajan, T.; Dwivedi, H.; Kumar, S.; Subramaniam, JR. Reserpine can confer stress tolerance and lifespan extension in the nematode C. elegans. Biogerontology 2008, 9, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Arya, U.; Dwivedi, H.; Subramaniam, J.R. Reserpine ameliorates Abeta toxicity in the Alzheimer’s disease model in Caenorhabditis elegans. Exp. Gerontol. 2009, 44, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.G.; Rogina, B.; Lavu, S.; Howitz, K.; Helfand, S.L.; Tatar, M.; Sinclair, D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 2004, 430, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Valenzano, D.R.; Terzibasi, E.; Genade, T.; Cattaneo, A.; Domenici, L.; Cellerino, A. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr. Biol. 2006, 16, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Rascón, B.; Hubbard, B.P.; Sinclair, D.A.; Amdam, G.V. The lifespan extension effects of resveratrol are conserved in the honey bee and may be driven by a mechanism related to caloric restriction. Aging 2012, 4, 499–508. [Google Scholar] [PubMed]
- Baur, J.A.; Pearson, K.J.; Price, N.L.; Jamieson, H.A.; Lerin, C.; Kalra, A.; Prabhu, V.V.; Allard, J.S.; Lopez-Lluch, G.; Lewis, K.; et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 2006, 444, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Maiuri, M.C.; Markaki, M.; Megalou, E.; Pasparaki, A.; Palikaras, K.; Criollo, A.; Galluzzi, L.; Malik, S.A.; Vitale, I.; et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010, 1, e10. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Minois, N.; Carmona-Gutierrez, D.; Bauer, M.A.; Rockenfeller, P.; Eisenberg, T.; Brandhorst, S.; Sigrist, S.J.; Kroemer, G.; Madeo, F. Spermidine promotes stress resistance in Drosophila melanogaster through autophagy-dependent and-independent pathways. Cell Death Dis. 2012, 3, e401. [Google Scholar] [CrossRef] [PubMed]
- Minois, N.; Rockenfeller, P.; Smith, T.K.; Carmona-Gutierrez, D. Spermidine feeding decreases age-related locomotor activity loss and induces changes in lipid composition. PLoS ONE 2014, 9, e102435. [Google Scholar] [CrossRef] [PubMed]
- Saul, N.; Pietsch, K.; Menzel, R.; Stürzenbaum, S.R.; Steinberg, C.E. The longevity effect of tannic acid in Caenorhabditis elegans: Disposable Soma meets hormesis. J. Gerontol. A Biol. Sci. Med. Sci. 2010, 65, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Cañuelo, A.; Gilbert-López, B.; Pacheco-Liñán, P.; Martínez-Lara, E.; Siles, E.; Miranda-Vizuete, A. Tyrosol, a main phenol present in extra virgin olive oil, increases lifespan and stress resistance in Caenorhabditis elegans. Mech. Ageing Dev. 2012, 133, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Strong, R.; Miller, R.A.; Astle, C.M.; Baur, J.A.; de Cabo, R.; Fernandez, E.; Guo, W.; Javors, M.; Kirkland, J.L.; Nelson, J.F.; et al. Evaluation of resveratrol, green tea extract, curcumin, oxaloacetic acid, and medium-chain triglyceride oil on life span of genetically heterogeneous mice. J. Gerontol. A Biol. Sci. Med. Sci. 2013, 68, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span-from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.A.; Richard, V.R.; Kyryakov, P.; Bourque, S.D.; Beach, A.; Burstein, M.T.; Glebov, A.; Koupaki, O.; Boukh-Viner, T.; Gregg, C.; et al. Chemical genetic screen identifies lithocholic acid as an anti-aging compound that extends yeast chronological life span in a TOR-independent manner, by modulating housekeeping longevity assurance processes. Aging 2010, 2, 393–414. [Google Scholar] [PubMed]
- Kaeberlein, M. Lessons on longevity from budding yeast. Nature 2010, 464, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Kapahi, P; Chen, D.; Rogers, A.N.; Katewa, S.D.; Li, P.W.; Thomas, E.L.; Kockel, L. With TOR, less is more: A key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010, 11, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Leonov, A.; Titorenko, V.I. A network of interorganellar communications underlies cellular aging. IUBMB Life 2013, 65, 665–674. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Arlia-Ciommo, A.; Leonov, A.; Piano, A.; Svistkova, V.; Titorenko, V.I. Cell-autonomous mechanisms of chronological aging in the yeast Saccharomyces cerevisiae. Microbial Cell 2014, 1, 164–178. [Google Scholar] [CrossRef]
- Arlia-Ciommo, A.; Piano, A.; Leonov, A.; Svistkova, V.; Titorenko, V.I. Quasi-programmed aging of budding yeast: A trade-off between programmed processes of cell proliferation, differentiation, stress response, survival and death defines yeast lifespan. Cell Cycle 2014, 13, 3336–3349. [Google Scholar] [CrossRef] [PubMed]
- Guarente, L. Aging research-where do we stand and where are we going. Cell 2014, 159, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, B.K.; Berger, S.L.; Brunet, A.; Campisi, J.; Cuervo, A.M.; Epel, E.S.; Franceschi, C.; Lithgow, G.J.; Morimoto, R.I.; Pessin, J.E.; et al. Geroscience: Linking aging to chronic disease. Cell 2014, 159, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Labbadia, J.; Morimoto, R.I. Proteostasis and longevity: When does aging really begin. F1000Prime Rep. 2014, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Partridge, L. Intervening in ageing to prevent the diseases of ageing. Trends Endocrinol. Metab. 2014, 25, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Harel, I.; Benayoun, B.A.; Machado, B.; Singh, P.P.; Hu, C.K.; Pech, M.F.; Valenzano, D.R.; Zhang, E.; Sharp, S.C.; Artandi, S.E.; et al. A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. Cell 2015, 160, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Hwang, W.; Artan, M.; Jeong, D.E.; Lee, S.J. Effects of nutritional components on aging. Aging Cell 2015, 14, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Piano, A.; Titorenko, V.I. The intricate interplay between mechanisms underlying aging and cancer. Aging Dis. 2015, 6, 56–75. [Google Scholar] [CrossRef] [PubMed]
- Southam, C.M.; Ehrlich, J. Effects of extract of western red-cedar heartwood on certain wood-decaying fungi in culture. Phytopathology 1943, 33, 517–524. [Google Scholar]
- Calabrese, E.J.; Baldwin, L.A. Defining hormesis. Hum. Exp. Toxicol. 2002, 21, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P. Hormesis defined. Ageing Res. Rev. 2008, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rattan, S.I. Hormesis in aging. Ageing Res. Rev. 2008, 7, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Bachmann, K.A.; Bailer, A.J.; Bolger, P.M.; Borak, J.; Cai, L.; Cedergreen, N.; Cherian, M.G.; Chiueh, C.C.; Clarkson, T.W.; et al. Biological stress response terminology: Integrating the concepts of adaptive response and preconditioning stress within a hormetic dose-response framework. Toxicol. Appl. Pharmacol. 2007, 222, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Mattson, M.P. Hormesis provides a generalized quantitative estimate of biological plasticity. J. Cell Commun. Signal. 2011, 5, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J. Paradigm lost, paradigm found: The re-emergence of hormesis as a fundamental dose response model in the toxicological sciences. Environ. Pollut. 2005, 138, 379–411. [Google Scholar] [CrossRef] [PubMed]
- Lamming, D.W.; Wood, J.G.; Sinclair, D.A. Small molecules that regulate lifespan: Evidence for xenohormesis. Mol. Microbiol. 2004, 53, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Howitz, K.T.; Sinclair, D.A. Xenohormesis: Sensing the chemical cues of other species. Cell 2008, 133, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, B.; Milbrandt, J. Resveratrol stimulates AMP kinase activity in neurons. Proc. Natl. Acad. Sci. USA 2007, 104, 7217–7222. [Google Scholar] [CrossRef] [PubMed]
- Armour, S.M.; Baur, J.A.; Hsieh, S.N.; Land-Bracha, A.; Thomas, S.M.; Sinclair, D.A. Inhibition of mammalian S6 kinase by resveratrol suppresses autophagy. Aging 2009, 1, 515–528. [Google Scholar] [PubMed]
- Blagosklonny, M.V. Inhibition of S6K by resveratrol: In search of the purpose. Aging 2009, 1, 511–514. [Google Scholar] [PubMed]
- Demidenko, Z.N.; Blagosklonny, M.V. At concentrations that inhibit mTOR, resveratrol suppresses cellular senescence. Cell Cycle 2009, 8, 1901–1904. [Google Scholar] [CrossRef] [PubMed]
- Blagosklonny, M.V. Rapamycin and quasi-programmed aging: Four years later. Cell Cycle 2010, 9, 1859–1862. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.A.; Kyryakov, P.; Bourque, S.D.; Titorenko, V.I. Xenohormetic, hormetic and cytostatic selective forces driving longevity at the ecosystemic level. Aging 2010, 2, 361–370. [Google Scholar] [PubMed]
- Burstein, M.T.; Beach, A.; Richard, V.R.; Koupaki, O.; Gomez-Perez, A.; Goldberg, A.A.; Kyryakov, P.; Bourque, S.D.; Glebov, A.; Titorenko, V.I. Interspecies chemical signals released into the environment may create xenohormetic, hormetic and cytostatic selective forces that drive the ecosystemic evolution of longevity regulation mechanisms. Dose-Response 2012, 10, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Vigh, L.; Horvàth, I.; van Hasselt, P.R.; Kuiper, P.J. Effect of frost hardening on lipid and fatty acid composition of chloroplast thylakoid membranes in two wheat varieties of contrasting hardiness. Plant Physiol. 1985, 79, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.J.; Joo, F.; Vigh, L. The role of unsaturated lipids in membrane structure and stability. Prog. Biophys. Mol. Biol. 1989, 53, 71–103. [Google Scholar] [CrossRef] [PubMed]
- Balogh, G.; Horváth, I.; Nagy, E.; Hoyk, Z.; Benkõ, S.; Bensaude, O.; Vígh, L. The hyperfluidization of mammalian cell membranes acts as a signal to initiate the heat shock protein response. FEBS J. 2005, 272, 6077–6086. [Google Scholar] [CrossRef] [PubMed]
- Nagy, E.; Balogi, Z.; Gombos, I.; Akerfelt, M.; Björkbom, A.; Balogh, G.; Török, Z.; Maslyanko, A.; Fiszer-Kierzkowska, A.; Lisowska, K.; et al. Hyperfluidization-coupled membrane microdomain reorganization is linked to activation of the heat shock response in a murine melanoma cell line. Proc. Natl. Acad. Sci. USA 2007, 104, 7945–7950. [Google Scholar] [CrossRef] [PubMed]
- Libertini, G. An adaptive theory of increasing mortality with increasing chronological age in populations in the wild. J. Theor. Biol. 1988, 132, 145–162. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, T.B. Comparative life spans of species: Why do species have the life spans they do. Am. J. Clin. Nutr. 1992, 55, 1191S–1195S. [Google Scholar] [PubMed]
- Skulachev, V.P. Aging is a specific biological function rather than the result of a disorder in complex living systems: Biochemical evidence in support of Weismann's hypothesis. Biochemistry 1997, 62, 1191–1195. [Google Scholar] [PubMed]
- Longo, V.D.; Mitteldorf, J.; Skulachev, V.P. Programmed and altruistic ageing. Nat. Rev. Genet. 2005, 6, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, T.C. Aging, evolvability, and the individual benefit requirement; medical implications of aging theory controversies. J. Theor. Biol. 2008, 252, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Ljubuncic, P.; Reznick, A.Z. The evolutionary theories of aging revisited—A mini-review. Gerontology 2009, 55, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Mitteldorf, J. Aging is not a process of wear and tear. Rejuvenation Res. 2010, 13, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, T.C. On the programmed/non-programmed aging controversy. Biochemistry 2012, 77, 729–732. [Google Scholar] [PubMed]
- Mitteldorf, J. Adaptive aging in the context of evolutionary theory. Biochemistry 2012, 77, 716–725. [Google Scholar] [PubMed]
- Trindade, L.S.; Aigaki, T.; Peixoto, A.A.; Balduino, A.; Mânica da Cruz, I.B.; Heddle, J.G. A novel classification system for evolutionary aging theories. Front. Genet. 2013, 4, 25. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leonov, A.; Arlia-Ciommo, A.; Piano, A.; Svistkova, V.; Lutchman, V.; Medkour, Y.; Titorenko, V.I. Longevity Extension by Phytochemicals. Molecules 2015, 20, 6544-6572. https://doi.org/10.3390/molecules20046544
Leonov A, Arlia-Ciommo A, Piano A, Svistkova V, Lutchman V, Medkour Y, Titorenko VI. Longevity Extension by Phytochemicals. Molecules. 2015; 20(4):6544-6572. https://doi.org/10.3390/molecules20046544
Chicago/Turabian StyleLeonov, Anna, Anthony Arlia-Ciommo, Amanda Piano, Veronika Svistkova, Vicky Lutchman, Younes Medkour, and Vladimir I. Titorenko. 2015. "Longevity Extension by Phytochemicals" Molecules 20, no. 4: 6544-6572. https://doi.org/10.3390/molecules20046544
APA StyleLeonov, A., Arlia-Ciommo, A., Piano, A., Svistkova, V., Lutchman, V., Medkour, Y., & Titorenko, V. I. (2015). Longevity Extension by Phytochemicals. Molecules, 20(4), 6544-6572. https://doi.org/10.3390/molecules20046544