Exploration of Scaffolds from Natural Products with Antiplasmodial Activities, Currently Registered Antimalarial Drugs and Public Malarial Screen Data
Abstract
:1. Introduction

2. Results and Discussion
2.1. Scaffold Diversity
2.1.1. Higher Scaffold Diversity Observed for the NAA Dataset
Scaffold Counts and Cumulative Scaffold Frequency Plots (CSFP)
| Ns/M | Nss/M | Nss/Ns | P25 | P50 | P75 | AUC | |
|---|---|---|---|---|---|---|---|
| Currently registered Antimalarial Drugs (CRAD) | 0.59 | 0.48 | 0.81 | 6.47 | 17.97 | 49.91 | 6794 |
| Natural products with in vitro antiplasmodial activity (NAA) | 0.29 | 0.17 | 0.57 | 1.65 | 6.75 | 27.58 | 8017 |
| Malarial screen data from Medicines for Malaria Venture (MMV) | 0.11 | 0.05 | 0.53 | 0.11 | 1.02 | 9.39 | 9043 |

2.1.2. Bioactivity Subgroups of NAA (HA, A, MA and LA)
Scaffold Counts and Cumulative Scaffold Frequency Plots (CSFP)
| Ns/M | Nss/M | Nss/Ns | P25 | P50 | P75 | AUC | |
|---|---|---|---|---|---|---|---|
| CRAD | 0.70 | 0.55 | 0.79 | 10.22 | 24.57 | 59.03 | 6247.6 |
| HA | 0.66 | 0.49 | 0.74 | 7.35 | 20.71 | 58.31 | 6410.2 |
| A | 0.60 | 0.40 | 0.67 | 6.59 | 18.96 | 54.50 | 6588.4 |
| MA | 0.64 | 0.49 | 0.77 | 6.16 | 18.53 | 55.68 | 6577.8 |
| LA | 0.52 | 0.38 | 0.73 | 4.19 | 13.96 | 46.50 | 7026.2 |

2.2. Molecular Similarity of Scaffolds Reveal Differences between the NAA and CRAD Datasets




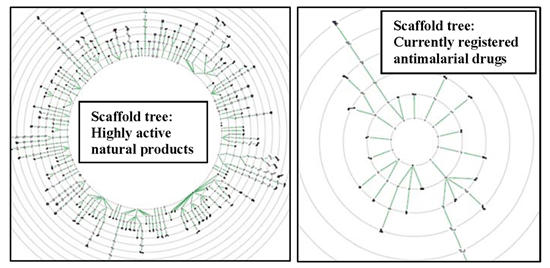
2.3. Scaffold Tree: Revealing Virtual Scaffolds

3. Materials and Methods
3.1. Datasets for Scaffold Analysis
3.2. Generation of Scaffolds and Scaffold Trees
3.3. Scaffold Diversity Analysis
3.3.1. Scaffold Counts
3.3.2. Cumulative Scaffold Frequency Plots (CSFP)
3.4. Molecular Similarity amongst Scaffolds
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lipkus, A.H.; Yuan, Q.; Lucas, K.A.; Funk, S.A.; Bartelt, W.F., III; Schenk, R.J.; Trippe, A.J. Structural diversity of organic chemistry. A scaffold analysis of the CAS registry. J. Org. Chem. 2008, 73, 4443–4451. [Google Scholar] [PubMed]
- Rates, S. Plants as source of drugs. Toxicon 2001, 39, 603–613. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M.; Snader, K.M. Natural products as sources of new drugs over the period 1981–2002. J. Nat. Prod. 2003, 66, 1022–1037. [Google Scholar] [CrossRef] [PubMed]
- Koehn, F.E.; Carter, G.T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed]
- Soh, P.N.; Benoit-Vical, F. Are West African plants a source of future antimalarial drugs? J. Ethnopharmacol. 2007, 114, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Frederich, M; Tits, M.; Angenot, L. Potential antimalarial activity of indole alkaloids. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 11–19. [Google Scholar]
- Ngueira, C.R.; Lopes, L.M. Antiplasmodial natural products. Molecules 2011, 16, 2146–2190. [Google Scholar] [CrossRef]
- Bon, R.S.; Waldmann, H. Bioactivity-guided navigation of chemical space. Acc. Chem. Res. 2010, 43, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Schneider, G. Scaffold architecture and pharmacophoric properties of natural products and trade drugs: Application in the design of natural product-based combinatorial libraries. J. Comb. Chem. 2001, 3, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Clardy, J.; Walsh, C. Lessons from natural molecules. Nature 2004, 432, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Boldi, A.M. Libraries from natural product-like scaffolds. Curr. Opin. Chem. Biol. 2004, 8, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Buss, A.D. Natural products—The future scaffolds for novel antibiotics? Biochem. Pharmacol. 2006, 71, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, K.; Baringhaus, K.; Schneider, G. Scaffold diversity of natural products: Inspiration for combinatorial library design. Nat. Prod. Rep. 2008, 25, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Morton, D.; Leach, S.; Cordier, C.; Warriner, S.; Nelson, A. Synthesis of Natural-Product-Like molecules with over eighty distinct scaffolds. Angew. Chem. Int. Ed. 2009, 48, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Welsch, M.E.; Snyder, S.A.; Stockwell, B.R. Privileged scaffolds for library design and drug discovery. Curr. Opin. Chem. Biol. 2010, 14, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Villar, H.O.; Hansen, M.R. Design of chemical libraries for screening. Expert Opin. Drug Discov. 2009, 4, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Willett, P. Chemoinformatics—Similarity and diversity in chemical libraries. Curr. Opin. Biotechnol. 2000, 11, 85–88. [Google Scholar] [CrossRef]
- Ertl, P.; Jelfs, S.; Mühlbacher, J.; Schuffenhauer, A.; Selzer, P. Quest for the rings. In silico exploration of ring universe to identify novel bioactive heteroaromatic scaffolds. J. Med. Chem. 2006, 49, 4568–4573. [Google Scholar] [PubMed]
- Kho, R.; Hodges, J.A.; Hansen, M.R.; Villar, H.O. Ring systems in mutagenicity databases. J. Med. Chem. 2005, 48, 6671–6678. [Google Scholar] [CrossRef] [PubMed]
- Krier, M.; Bret, G.; Rognan, D. Assessing the scaffold diversity of screening libraries. J. Chem. Inf. Model. 2006, 46, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Bemis, G.W.; Murcko, M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996, 39, 2887–2893. [Google Scholar] [CrossRef] [PubMed]
- Shelat, A.A.; Guy, R.K. Scaffold composition and biological relevance of screening libraries. Nat. Chem. Biol. 2007, 3, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Langdon, S.R.; Brown, N.; Blagg, J. Scaffold diversity of exemplified medicinal chemistry space. J. Chem. Inf. Model. 2011, 51, 2174–2185. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wassermann, A.M.; Lounkine, E.; Bajorath, J. Systematic analysis of public domain compound potency data identifies selective molecular scaffolds across druggable target families. J. Med. Chem. 2009, 53, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Lameijer, E.; Kok, J.N.; Bäck, T.; IJzerman, A.P. Mining a chemical database for fragment co-occurrence: Discovery of “chemical cliches”. J. Chem. Inf. Model. 2006, 46, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, K.; Schneider, G. Properties and architecture of drugs and natural products revisited. Curr. Chem. Biol. 2007, 1, 115–127. [Google Scholar] [CrossRef]
- Wetzel, S.; Schuffenhauer, A.; Roggo, S.; Ertl, P.; Waldmann, H. Cheminformatic analysis of natural products and their chemical space. CHIMIA 2007, 61, 355–360. [Google Scholar] [CrossRef]
- Wetzel, S.; Klein, K.; Renner, S.; Roah, D.; Oprea, T.I.; Mutzel, P.; Waldman, H. Interactive exploration of chemical space with scaffold hunter. Nat. Chem. Biol. 2009, 5, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Galloway, W.R.; Wilcke, D.; Nie, F.; Hadje-Georgiou, K.; Laraia, L.; Spring, D.R. Diversity-oriented synthesis: Developing new chemical tools to probe and modulate biological systems. In Concepts and Case Studies in Chemical Biology; Waldmann, H., Janning, P., Eds.; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2014; pp. 379–390. [Google Scholar]
- Lipinski, C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Li, Y.; Wang, J.; Xu, X.; Xu, L.; Wang, X.; Chen, L.; Hou, T. Drug-likeness analysis of traditional chinese medicines: 2. Characterization of scaffold architectures for drug-like compounds, non-drug-like compounds, and natural compounds from traditional chinese medicines. J. Cheminform. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Ortholand, J.; Ganesan, A. Natural products and combinatorial chemistry: Back to the future. Curr. Opin. Chem. Biol. 2004, 8, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Galloway, W.R.; Isidro-Llobet, A.; Spring, D.R. Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat. Commun. 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Guha, R.; Giulianotti, M.A.; Pinilla, C.; Houghten, R.A.; Medina-Franco, J.L. Chemoinformatic analysis of combinatorial libraries, drugs, natural products, and molecular libraries small molecule repository. J. Chem. Inf. Model. 2009, 49, 1010–1024. [Google Scholar] [CrossRef] [PubMed]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kotter, T.; Meinl, T.; Ohl, P.; Thiel, K.; Wiswedel, B. KNIME-the konstanz information miner: Version 2.0 and beyond. SIGKDD Explor. 2009, 11, 26–31. [Google Scholar] [CrossRef]
- Landrum, G. RDKit: Open-Source Cheminformatics. Available online: http://www.rdkit.org (accessed on 24 September 2015).
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program for Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Egieyeh, S.; Syce, J.; Christoffels, A.; Malan, S.F. Exploration of Scaffolds from Natural Products with Antiplasmodial Activities, Currently Registered Antimalarial Drugs and Public Malarial Screen Data. Molecules 2016, 21, 104. https://doi.org/10.3390/molecules21010104
Egieyeh S, Syce J, Christoffels A, Malan SF. Exploration of Scaffolds from Natural Products with Antiplasmodial Activities, Currently Registered Antimalarial Drugs and Public Malarial Screen Data. Molecules. 2016; 21(1):104. https://doi.org/10.3390/molecules21010104
Chicago/Turabian StyleEgieyeh, Samuel, James Syce, Alan Christoffels, and Sarel F. Malan. 2016. "Exploration of Scaffolds from Natural Products with Antiplasmodial Activities, Currently Registered Antimalarial Drugs and Public Malarial Screen Data" Molecules 21, no. 1: 104. https://doi.org/10.3390/molecules21010104
APA StyleEgieyeh, S., Syce, J., Christoffels, A., & Malan, S. F. (2016). Exploration of Scaffolds from Natural Products with Antiplasmodial Activities, Currently Registered Antimalarial Drugs and Public Malarial Screen Data. Molecules, 21(1), 104. https://doi.org/10.3390/molecules21010104