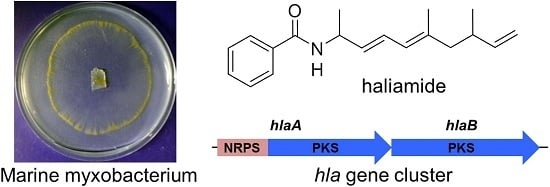
Isolation and Biosynthetic Analysis of Haliamide, a New PKS-NRPS Hybrid Metabolite from the Marine Myxobacterium Haliangium ochraceum
Abstract
:1. Introduction

2. Results and Discussion
2.1. Isolation and Structural Elucidation
| Position | δC | δH mult. (J in Hz) |
|---|---|---|
| 1 | 20.8 | 1.38 d (6.8) |
| 2 | 47.0 | 4.84 ddq (6.0, 7.2, 6.8) |
| 3 | 131.9 | 5.63 dd (6.0, 15.2) |
| 4 | 126.8 | 6.46 dd (10.8, 15.2) |
| 5 | 125.5 | 5.81 d (10.8) |
| 6 | 138.0 | - |
| 7 | 47.4 | 1.97 dd (7.6, 13.6), 2.10 dd (7.2, 13.6) |
| 8 | 35.6 | 2.36 dddq (6.8, 7.2, 7.6, 6.4) |
| 9 | 144.2 | 5.73 ddd (6.8, 10.4, 17.2) |
| 10 | 112.4 | 4.91 d (10.4), 4.96 d (17.2) |
| 11 | 19.5 | 0.95 d (6.4) |
| 12 | 16.7 | 1.73 s |
| 1′ | 166.5 | - |
| 2′ | 134.8 | - |
| 3′, 7′ | 126.8 | 7.77 d (7.5) |
| 4′,6′ | 128.5 | 7.43 t (7.5) |
| 5′ | 131.4 | 7.49 t (7.5) |
| NH | - | 6.04 d (7.2) |

2.2. Identification of the Biosynthetic Precursors

2.3. Proposed Biosynthetic Mechanism of Haliamide (1)

2.4. Bioactivities of Haliamide (1)
3. Materials and Methods
3.1. General
3.2. Isolation
3.3. Feeding Experiments with Stable Isotope Labeled Precursors
3.4. Analysis of the Haliamide Biosynthetic Gene Cluster (hla)
3.5. Bioassay
3.5.1. Cytotoxicity Assay
3.5.2. Anti-Oomycete Activity
3.5.3. MIC Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Weissman, K.J.; Mueller, R. A brief tour of myxobacterial secondary metabolism. Bioorg. Med. Chem. 2009, 17, 2121–2136. [Google Scholar] [CrossRef] [PubMed]
- Reichenbach, H. Myxobacteria, producers of novel bioactive substances. J. Ind. Microbiol. Biotechnol. 2001, 27, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, S.C.; Muller, R. Myxobacteria—“Microbial factories” for the production of bioactive secondary metabolites. Mol. Biosyst. 2009, 5, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, T.; Jojima, Y.; Fudou, R.; Tokura, M.; Hiraishi, A.; Yamanaka, S. Enhygromyxa salina gen. nov., sp nov., a slightly halophilic myxobacterium isolated from the coastal areas of Japan. Syst. Appl. Microbiol. 2003, 26, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Schaeberle, T.F.; Goralski, E.; Neu, E.; Erol, O.; Hoelzl, G.; Doermann, P.; Bierbaum, G.; Koenig, G.M. Marine Myxobacteria as a Source of Antibiotics—Comparison of Physiology, Polyketide-Type Genes and Antibiotic Production of Three New Isolates of Enhygromyxa salina. Mar. Drugs 2010, 8, 2466–2479. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, T.; Jojima, Y.; Fudou, R.; Hiraishi, A.; Ahn, J.W.; Yamanaka, S. Plesiocystis pacifica gen. nov., sp nov., a marine myxobacterium that contains dihydrogenated menaquinone, isolated from the Pacific coasts of Japan. Int. J. Syst. Evol. Microbiol. 2003, 53, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, T.; Jojima, Y.; Hayakawa, A.; Fujii, T.; Yamanaka, S.; Fudou, R. Pseudenhygromyxa salsuginis gen. nov., sp nov., a myxobacterium isolated from an estuarine marsh. Int. J. Syst. Evol. Microbiol. 2013, 63, 1360–1369. [Google Scholar] [CrossRef] [PubMed]
- Still, P.C.; Johnson, T.A.; Theodore, C.M.; Loveridge, S.T.; Crews, P. Scrutinizing the Scaffolds of Marine Biosynthetics from Different Source Organisms: Gram-Negative Cultured Bacterial Products Enter Center Stage. J. Nat. Prod. 2014, 77, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Fudou, R.; Iizuka, T.; Nakajima, D.; Okazaki, K.; Shibata, D.; Ojika, M.; Harayama, S. PCR Detection of Type I Polyketide Synthase Genes in Myxobacteria. Appl. Environ. Microbiol. 2008, 74, 5571–5574. [Google Scholar] [CrossRef] [PubMed]
- Fudou, R.; Iizuka, T.; Yamanaka, S. Haliangicin, a novel antifungal metabolite produced by a marine myxobacterium 1. Fermentation and biological characteristics. J. Antibiot. 2001, 54, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Fudou, R.; Iizuka, T.; Sato, S.; Ando, T.; Shimba, N.; Yamanaka, S. Haliangicin, a novel antifungal metabolite produced by a marine myxobacterium 2. Isolation and structural elucidation. J. Antibiot. 2001, 54, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Kundim, B.A.; Itou, Y.; Sakagami, Y.; Fudou, R.; Iizuka, T.; Yamanaka, S.; Ojika, M. New haliangicin isomers, potent antifungal metabolites produced by a marine myxobacterium. J. Antibiot. 2003, 56, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Ojika, M.; Inukai, Y.; Kito, Y.; Hirata, M.; Iizuka, T.; Fudou, R. Miuraenamides: Antimicrobial cyclic depsipeptides isolated from a rare and slightly halophilic myxobacterium. Chem. Asian J. 2008, 3, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, T.; Fudou, R.; Jojima, Y.; Ogawa, S.; Yamanaka, S.; Inukai, Y.; Ojika, M. Miuraenamides A and B, novel antimicrobial cyclic depsipeptides from a new slightly halophilic myxobacterium: Taxonomy, production, and biological properties. J. Antibiot. 2006, 59, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Felder, S.; Dreisigacker, S.; Kehraus, S.; Neu, E.; Bierbaum, G.; Wright, P.R.; Menche, D.; Schaberle, T.F.; Konig, G.M. Salimabromide: Unexpected Chemistry from the Obligate Marine Myxobacterium Enhygromyxa salina. Chem. Eur. J. 2013, 19, 9319–9324. [Google Scholar] [CrossRef] [PubMed]
- Felder, S.; Kehraus, S.; Neu, E.; Bierbaum, G.; Schaberle, T.F.; Konig, G.M. Salimyxins and Enhygrolides: Antibiotic, Sponge-Related Metabolites from the Obligate Marine Myxobacterium Enhygromyxa salina. ChemBioChem 2013, 14, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Ligon, J.; Hill, S.; Beck, J.; Zirkle, R.; Molnar, I.; Zawodny, J.; Money, S.; Schupp, T. Characterization of the biosynthetic gene cluster for the antifungal polyketide soraphen A from Sorangium cellulosum So ce26. Gene 2002, 285, 257–267. [Google Scholar] [CrossRef]
- Kalaitzis, J.A.; Cheng, Q.; Meluzzi, D.; Xiang, L.K.; Izumikawa, M.; Dorrestein, P.C.; Moore, B.S. Policing starter unit selection of the enterocin type II polyketide synthase by the type II thioesterase EncL. Bioorg. Med. Chem. 2011, 19, 6633–6638. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. antiSMASH 2.0-a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013, 41, W204–W212. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Plaza, A.; Dubiella, C.; Groll, M.; Kaiser, M.; Muller, R. Macyranones: Structure, Biosynthesis, and Binding Mode of an Unprecedented Epoxyketone that Targets the 20S Proteasome. J. Am. Chem. Soc. 2015, 137, 8121–8130. [Google Scholar] [CrossRef] [PubMed]
- Yadav, G.; Gokhale, R.S.; Mohanty, B. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 2003, 328, 335–363. [Google Scholar] [CrossRef]
- Petkovic, H.; Sandmann, A.; Challis, L.R.; Hecht, H.J.; Silakowski, B.; Low, L.; Beeston, N.; Kuscer, E.; Garcia-Bernardo, J.; Leadlay, P.F.; et al. Substrate specificity of the acyl transferase domains of EpoC from the epothilone polyketide synthase. Org. Biomol. Chem. 2008, 6, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.X.; Sitachitta, N.; Rossi, J.V.; Roberts, M.A.; Flatt, P.M.; Jia, J.Y.; Sherman, D.H.; Gerwick, W.H. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2004, 67, 1356–1367. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.C.; Wang, B.; Kulkarni, A.; Gehret, J.J.; Lloyd, K.R.; Gerwick, L.; Gerwick, W.H.; Wipf, P.; Hakansson, K.; Smith, J.L.; et al. Polyketide Decarboxylative Chain Termination Preceded by O-Sulfonation in Curacin A Biosynthesis. J. Am. Chem. Soc. 2009, 131, 16033–16035. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, N.; Daum, C.; Lang, E.; Abt, B.; Kopitz, M.; Saunders, E.; Lapidus, A.; Lucas, S.; del Rio, T.G.; Nolan, M.; et al. Complete genome sequence of Haliangium ochraceum type strain (SMP-2(T)). Stand. Genom. Sci. 2010, 2, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.N.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; de Weese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Sample Availability: Sample of the compound 1 is available from the authors.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Tomura, T.; Sato, J.; Iizuka, T.; Fudou, R.; Ojika, M. Isolation and Biosynthetic Analysis of Haliamide, a New PKS-NRPS Hybrid Metabolite from the Marine Myxobacterium Haliangium ochraceum. Molecules 2016, 21, 59. https://doi.org/10.3390/molecules21010059
Sun Y, Tomura T, Sato J, Iizuka T, Fudou R, Ojika M. Isolation and Biosynthetic Analysis of Haliamide, a New PKS-NRPS Hybrid Metabolite from the Marine Myxobacterium Haliangium ochraceum. Molecules. 2016; 21(1):59. https://doi.org/10.3390/molecules21010059
Chicago/Turabian StyleSun, Yuwei, Tomohiko Tomura, Junichi Sato, Takashi Iizuka, Ryosuke Fudou, and Makoto Ojika. 2016. "Isolation and Biosynthetic Analysis of Haliamide, a New PKS-NRPS Hybrid Metabolite from the Marine Myxobacterium Haliangium ochraceum" Molecules 21, no. 1: 59. https://doi.org/10.3390/molecules21010059
APA StyleSun, Y., Tomura, T., Sato, J., Iizuka, T., Fudou, R., & Ojika, M. (2016). Isolation and Biosynthetic Analysis of Haliamide, a New PKS-NRPS Hybrid Metabolite from the Marine Myxobacterium Haliangium ochraceum. Molecules, 21(1), 59. https://doi.org/10.3390/molecules21010059