Multicomponent Analysis of the Differential Induction of Secondary Metabolite Profiles in Fungal Endophytes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Fungal Isolation and Identification
2.2. Screening of Epigenetic Modifiers Effects on Fungal Endophytes
2.3. Standardization of Cultivation Conditions of Dothiora sp.
2.3.1. Effects of Epigenetic Modifiers on Dothiora sp. Submerged Culturing
2.3.2. Metabolomic Evaluation of the Changes in the SM Profile of Dothiora sp. by Epigenetic Modifiers
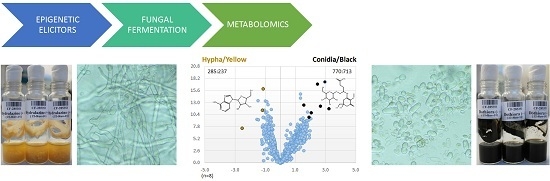
2.3.3. Identification of Molecules Produced in the Fermentations with Modifiers
2.3.4. Dothiora sp. Biomarkers Related to Growth Morphology
3. Experimental Section
3.1. Isolation Cultures and Characterization
3.2. Epigenetic Modifiers Stock Solutions
3.3. Inocula Preparation
3.4. Production Fermentations
3.5. Chemical Extraction
3.6. UHPLC-UV Profile Analysis
3.7. LC/MS Data and Metabolomics Evaluation
3.8. UHPLC-HRMS Database Matching of Known Metabolites and Antibiotics
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Xiang, S.; Liang-Dong, G. Endophytic fungal diversity: Review of traditional and molecular techniques. Mycology 2002, 3, 65–76. [Google Scholar]
- Cichewicz, R.H. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 2010, 27, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Brosch, G.; Loidl, P.; Graessle, S. Histone modifications and chromatin dynamics: A focus on filamentous fungi. Fems Microbiol. Rev. 2008, 32, 409–439. [Google Scholar] [CrossRef] [PubMed]
- Preyat, N.; Leo, O. Sirtuin deacylases: A molecular link between metabolism and immunity. J. Leukoc. Biol. 2013, 93, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Avalos, J.L.; Bever, K.M.; Wolberger, C. Mechanism of sirtuin inhibition by nicotinamide: Altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol. Cell 2005, 18, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.M.; Bradshaw, E.; Seipke, R.F.; Hutchings, M.I.; McArthur, M. Use and discovery of chemical elicitors that stimulate biosynthetic gene clusters in Streptomyces bacteria. Methods Enzymol. 2012, 517, 367–385. [Google Scholar] [PubMed]
- Moskaug, J.O.; Carlsen, H.; Myhrstad, M.; Blomhoff, R. Molecular imaging of the biological effects of quercetin and quercetin-rich foods. Mech. Ageing Dev. 2004, 125, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Morita, S.; Taniguchi, T.; Monde, K.; Oshima, Y. Epigenetic stimulation of polyketide production in Chaetomium cancroideum by an NAD(+)-dependent HDAC inhibitor. Org. Biomol. Chem. 2015, 14, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, H.C.; Drašković, M.; Crews, P. Histone Deacetylase Inhibitors as a Tool to Up-regulate New Fungal Biosynthetic Products—Isolation of EGM-556, a Cyclodepsipeptide, from Microascus sp. Org. Lett. 2011, 13, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Beau, J.; Mahid, N.; Burda, W.N.; Harrington, L.; Shaw, L.N.; Mutka, T.; Kyle, D.E.; Barisic, B.; van Olphen, A.; Baker, B.J. Epigenetic Tailoring for the Production of Anti-Infective Cytosporones from the Marine Fungus Leucostoma persoonii. Mar. Drugs 2012, 10, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Awakawa, T.; Sun, J.Y.; Wakimoto, T.; Abe, I. Epigenetic modifier-induced biosynthesis of novel fusaric acid derivatives in endophytic fungi from Datura stramonium L. Nat. Prod. Bioprospect. 2013, 3, 20–23. [Google Scholar] [CrossRef]
- Albright, J.C.; Henke, M.T.; Soukup, A.A.; McClure, R.A.; Thomson, R.J.; Keller, N.P.; Kelleher, N.L. Large-scale metabolomics reveals a complex response of Aspergillus nidulans to epigenetic perturbation. ACS Chem. Biol. 2015, 10, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Hur, M.; Campbell, A.A.; Almeida-de-Macedo, M.; Li, L.; Ransom, N.; Jose, A.; Wurtele, E.S. A global approach to analysis and interpretation of metabolic data for plant natural product discovery. Nat. Prod. Rep. 2013, 30, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Bills, G.F.; González-Menéndez, V.; Platas, G. Kabatiella bupleuri sp. nov. (Dothideales), a pleomorphic epiphyte and endophyte of the Mediterranean plant Bupleurum gibraltarcum (Apiaceae). Mycologia. 2012, 104, 962–973. [Google Scholar] [CrossRef] [PubMed]
- DelaCruz, M.; Martín, J.; González-Menéndez, V.; Pérez-Victoria, I.; Moreno, C.; Tormo, J.R.; El Aouad, N.; Guarro, J.; Vicente, F.; Reyes, F.; et al. Chemical and physical modulation of antibiotic activity in emericella species. Chem. Biodivers. 2012, 9, 1095–1113. [Google Scholar] [CrossRef] [PubMed]
- Stanbury, P.F.; Whitakerm, A.; Hall, S.J. The isolation, preservation and improvement of industrially important microorganisms. In Principles of Fermentation Technology, 2nd ed.; Elsevier Science: Kidlington, UK, 1997; pp. 35–85. [Google Scholar]
- Dhandhukia, P.C.; Thakkar, V.R. Standardization of growth and fermentation criteria of Lasiodiplodia theobromae for production of jasmonic acid. J. Biotechnol. 2007, 6, 707–712. [Google Scholar]
- Leung, P.H.; Zhang, Q.X.; Wu, J.Y. Mycelium cultivation, chemical composition and antitumour activity of a Tolypocladium sp. fungus isolated from wild Cordyceps sinensis. J. Appl. Microbiol. 2006, 101, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Pirttilä, A.M.; Joensuu, P.; Pospiech, H.; Jalonen, J.; Hohtola, A. Bud endophytes of Scots pine produce adenine derivatives and other compounds that affect morphology and mitigate browning of callus cultures. Physiol. Plant. 2004, 121, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Gorbushina, A.A.; Whitehead, K.; Dornieden, T.; Niesse, A.; Schulte, A.; Hedges, J.I. Black fungal colonies as units of survival: Hyphal mycosporines synthesized by rock-dwelling microcolonial fungi. Can. J. Bot. 2003, 81, 131–138. [Google Scholar] [CrossRef]
- Leach, C.M. Ultraviolet-absorbing substances associated with light-induced sporulation in fungi. Can. J. Bot. 1965, 43, 185–200. [Google Scholar] [CrossRef]
- Trione, E.J.; Leach, C.M.; Mutch, J.T. Sporogenic substances isolated from fungi. Nature 1966, 212, 163–164. [Google Scholar] [CrossRef] [PubMed]
- Young, V.J.; Patterson, A. UV protective compound from Glomerella cingulate a mycosporine. Phytochemistry 1982, 21, 1075–1077. [Google Scholar] [CrossRef]
- Leite, B.; Nicholson, R.L. Mycosporine-alanine: A self-inhibitor of germination from the conidial mucilage of Colletotrichum graminicola. Exp. Mycol. 1992, 16, 76–86. [Google Scholar] [CrossRef]
- Sommaruga, R.; Libkind, D.; van Broock, M.; Whitehead, K. Mycosporine-glutaminol-glucoside, a UV-absorbing compound of two Rhodotorula yeast species. Yeast 2004, 21, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Carreto, J.I.; Carignan, M.O. Mycosporine-like amino acids: Relevant secondary metabolites. Chemical and ecological aspects. Mar Drugs. 2011, 9, 387–446. [Google Scholar] [CrossRef] [PubMed]
- Bills, G.F.; Platas, G.; Fillola, A.; Jiménez, M.R.; Collado, J.; Vicente, F.; Martín, J.; González, A.; Bur-Zimmermann, J.; Tormo, J.R.; et al. Enhancement of antibiotic and secondary metabolite detection from filamentous fungi by growth on nutritional arrays. J. Appl. Microbiol. 2008, 104, 1644–1658. [Google Scholar] [CrossRef] [PubMed]
- González-Menéndez, V.; Asensio, F.; Moreno, C.; de Pedro, N.; Monteiro, M.C.; de la Cruz, M.; Vicente, F.; Bills, G.F.; Reyes, F.; Genilloud, O.; et al. Assessing the effects of adsorptive polymeric resin additions on fungal secondary metabolite chemical diversity. Mycology 2014, 5, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Churchill, G.A. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 2003, 4, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sample Availability: Samples of the compounds 9–12 are available from the authors.








| Family | Strain Code | Species | Host Plant | Origin | Genbank |
|---|---|---|---|---|---|
| Dothideaceae | CF-277039 | Kabatiella bupleuri | Bupleurum gibraltarium | Presa de Quentar, (Granada) | JN886788 |
| Dothideaceae | CF-277101 | Selenophoma sp. | Spartium junceum | Fuente del Hervidero, (Granada) | JN886791 |
| Dothideaceae | CF-280549 | Kabatiella sp. | Bupleurum spinosum | Camino de los neveros (Granada) | KU295574 |
| Dothideaceae | CF-285353 | Dothiora sp. | Launaea arborescens | Tabernas (Almeria) | KU295575 |
| Dothideaceae | CF-285359 | Kabatiella sp. | Asparagus horridus | Tabernas (Almeria) | KU295576 |
| Dothideaceae | CF-285463 | Selenophoma juncea | Salsola oppositifolia | Tabernas (Almeria) | KU295578 |
| Dothideaceae | CF-285762 | Aureobasidium pullulans | Inula chrihtmoides | Punta Entinas (Almeria) | KU295579 |
| Chaetothyriaceae | CF-285360 | Knufia sp. | Asparagus horridus | Tabernas (Almeria) | KU295577 |
| Phaeosphaeriaceae | CF-285372 | Chaetosphaeronema sp. | Anabasis articulata | Tabernas (Almeria) | KU295581 |
| Planistromellaceae | CF-282001 | Loratospora sp. | Retama sphaerocarpa | Albuñuelas (Granada) | KU295580 |
| Sporormiaceae | CF-282341 | Preussia sp. | Dittrichia viscosa | Alhendin (Granada) | KU295582 |
| Sporormiaceae | CF-285375 | Preussia australis | Launaea arborescens | Tabernas (Almeria) | KU295583 |
| Xylariaceae | CF-285461 | Xylaria sp. | Thymelaea hirsuta | Tabernas (Almeria) | KU295584 |
| Strain ID | 5-Azacitidine (1) | Hydralazine (2) | Nicotinamide (3) | Quercetin (4) | SBHA (5) | Sodium Butyrate (6) | Valproic (7) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (− +) | (+ +) | (− +) | (+ +) | (− +) | (+ +) | (− +) | (+ +) | (− +) | (+ +) | (− +) | (+ +) | (− +) | (+ +) | |
| CF-277039 | - | - | - | - | - | - | - | - | p | m | pp | mp | m | m |
| CF-277101 | p | p | p | p | p | p | p | p | p | dmp | p | p | p | p |
| CF-280549 | m | m | m | - | m | - | m | m | m | mp | m | - | p | - |
| CF-285353 | - | ddmpp | - | ddmpp | - | - | - | ddmpp | p | m | p | - | - | m |
| CF-285359 | - | m | - | m | - | m | m | - | - | m | - | - | - | m |
| CF-285463 | - | mp | - | - | - | mp | - | mp | - | mp | - | mp | - | mp |
| CF-285762 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CF-285360 | - | - | - | - | - | p | - | - | - | p | - | p | - | - |
| CF-285372 | p | p | p | p | - | - | - | p | p | p | p | p | - | - |
| CF-282001 | pp | p | ppp | pp | - | - | p | dppp | p | pp | pd | ddpp | m | m |
| CF-282341 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CF-285375 | p | p | - | - | - | - | - | - | - | - | p | - | - | - |
| CF-285461 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Menéndez, V.; Pérez-Bonilla, M.; Pérez-Victoria, I.; Martín, J.; Muñoz, F.; Reyes, F.; Tormo, J.R.; Genilloud, O. Multicomponent Analysis of the Differential Induction of Secondary Metabolite Profiles in Fungal Endophytes. Molecules 2016, 21, 234. https://doi.org/10.3390/molecules21020234
González-Menéndez V, Pérez-Bonilla M, Pérez-Victoria I, Martín J, Muñoz F, Reyes F, Tormo JR, Genilloud O. Multicomponent Analysis of the Differential Induction of Secondary Metabolite Profiles in Fungal Endophytes. Molecules. 2016; 21(2):234. https://doi.org/10.3390/molecules21020234
Chicago/Turabian StyleGonzález-Menéndez, Víctor, Mercedes Pérez-Bonilla, Ignacio Pérez-Victoria, Jesús Martín, Francisca Muñoz, Fernando Reyes, José R. Tormo, and Olga Genilloud. 2016. "Multicomponent Analysis of the Differential Induction of Secondary Metabolite Profiles in Fungal Endophytes" Molecules 21, no. 2: 234. https://doi.org/10.3390/molecules21020234
APA StyleGonzález-Menéndez, V., Pérez-Bonilla, M., Pérez-Victoria, I., Martín, J., Muñoz, F., Reyes, F., Tormo, J. R., & Genilloud, O. (2016). Multicomponent Analysis of the Differential Induction of Secondary Metabolite Profiles in Fungal Endophytes. Molecules, 21(2), 234. https://doi.org/10.3390/molecules21020234