Design, Synthesis, Antimicrobial Evaluation and Molecular Modeling Study of 1,2,4-Triazole-Based 4-Thiazolidinones
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Studies
2.2.1. Antimicrobial Activity
2.2.2. Antibacterial Activity
2.2.3. Antifungal Activity
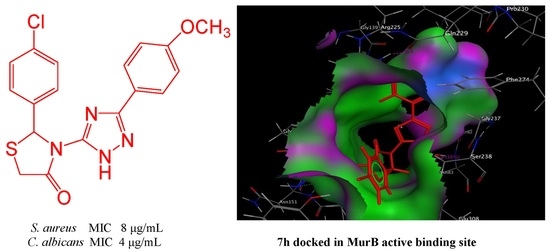
2.2.4. Molecular Modeling Study
3. Materials and Methods
3.1. General
3.1.1. General Method
3.1.2. General Method for Synthesis of Schiff Bases (6c–l)
3.1.3. General Method for the Synthesis of the Compounds (7c–l)
3.2. Biological Evaluation
3.2.1. Inhibition Zone Measurement Using the Cup Plate Method
3.2.2. Determination of MIC
3.3. Molecular Modeling Studies
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Carlet, J.; Collignon, P.; Goldmann, D.; Goossens, H.; Gyssens, I.C.; Harbarth, S.; Jarlier, V.; Levy, S.B.; N’Doye, B.; Pittet, D.; et al. Society’s failure to protect a precious resource: Antibiotics. Lancet 2011, 378, 369–371. [Google Scholar] [CrossRef]
- World Health Organization. The World Health Report 2000—Health Systems: Improving Performance; World Health Organization: Geneva, Switzerland, 2000. [Google Scholar]
- Mark, W.; Jeremy, F. Policy: An intergovernmental panel on antimicrobial resistance. Nature 2014, 509, 555–557. [Google Scholar]
- Anil, K.; Eric, A.; Nacer, L.; Jerome, G.; Koen, A. The challenge of new drug discovery for tuberculosis. Nature 2011, 469, 483–490. [Google Scholar]
- Rogers, H.J.; Perkins, H.R.; Ward, J.B. Biosynthesis of peptidoglycan. In Microbial Cell Walls and Membranes; Chapman & Hall: London, UK, 1980. [Google Scholar]
- Van Heijenoort, J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 2001, 18, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Mengin-Lecreulx, D.; Flouret, B.; van Heijenoort, J. Cytoplasmic steps of peptidoglycan synthesis in escherichia coli. J. Bacteriol. 1982, 151, 1109–1117. [Google Scholar] [PubMed]
- Andres, C.J.; Bronson, J.J.; D’Andrea, S.V.; Despande, M.S.; Falk, P.J.; Grant-young, K.A.; Harte, W.E.; Ho, H.T.; Misco, P.F.; Robertson, J.G.; et al. 4-Thiazolidinones: Novel inhibitors of the bacterial enzyme murB. Bioorg. Med. Chem. Lett. 2000, 10, 715–717. [Google Scholar] [CrossRef]
- Bronson, J.J.; DenBleyker, K.L.; Falk, P.J.; Mate, R.A.; Ho, H.T.; Pucci, M.J.; Snyder, L.B. Discovery of the first antibacterial small molecule inhibitors of murb. Bioorg. Med. Chem. Lett. 2003, 13, 873–875. [Google Scholar] [CrossRef]
- Jain, A.K.; Vaidya, A.; Ravichandran, V.; Kashaw, S.K.; Agrawal, R.K. Recent developments and biological activities of thiazolidinone derivatives: A review. Bioorg. Med. Chem. 2012, 20, 3378–3395. [Google Scholar] [CrossRef] [PubMed]
- Deep, A.; Jain, S.; Sharma, P.C.; Mittal, S.K.; Phogat, P.; Malhotra, M. Synthesis, characterization and antimicrobial evaluation of 2,5-disubstituted-4-thiazolidinone derivatives. Arab. J. Chem. 2014, 7, 287–291. [Google Scholar] [CrossRef]
- Tripathi, A.C.; Gupta, S.J.; Fatima, G.N.; Sonar, P.K.; Verma, A.; Saraf, S.K. 4-Thiazolidinones: The advances continue. Eur. J. Med. Chem. 2014, 72, 52–77. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Mishra, L.; Noolvi, M.; Karpoormath, R.; Cameotra, S.S. Synthesis, in vitro evaluation, and molecular docking studies of azetidinones and thiazolidinones of 2-amino-5-cyclopropyl-1,3,4-thiadiazole as antibacterial agents. Arch. Pharm. 2014, 347, 668–684. [Google Scholar] [CrossRef] [PubMed]
- Omar, K.; Geronikaki, A.; Zoumpoulakis, P.; Camoutsis, C.; Soković, M.; Ćirić, A.; Glamočlija, J. Novel 4-thiazolidinone derivatives as potential antifungal and antibacterial drugs. Bioorg. Med. Chem. 2010, 18, 426–432. [Google Scholar] [CrossRef] [PubMed]
- El Bialy, S.A.; Nagy, M.M.; Abdel-Rahman, H.M. Efficient regioselective three-component domino synthesis of 3-(1,2,4-triazol-5-yl)-1,3-thiazolidin-4-ones as potent antifungal and antituberculosis agents. Arch. Pharm. 2011, 344, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, H.M.; Hussein, M.A. Synthesis of beta-hydroxypropanoic acid derivatives as potential anti-inflammatory, analgesic and antimicrobial agents. Arch. Pharm. 2006, 339, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wen, Q.; Wang, S.F.; Shahla Karim, B.; Yang, Y.S.; Liu, J.J.; Zhang, W.M.; Zhu, H.L. Design, synthesis and antibacterial activities of 5-(pyrazin-2-yl)-4H-1,2,4-triazole-3-thiol derivatives containing schiff base formation as fabh inhibitory. Bioorg. Med. Chem. Lett. 2014, 24, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Romagnoli, R.; Baraldi, P.G.; Salvador, M.K.; Prencipe, F.; Bertolasi, V.; Cancellieri, M.; Brancale, A.; Hamel, E.; Castagliuolo, I.; Consolaro, F.; et al. Synthesis, antimitotic and antivascular activity of 1-(3′,4′,5′-trimethoxybenzoyl)-3-arylamino-5-amino-1,2,4-triazoles. J. Med. Chem. 2014, 57, 6795–6808. [Google Scholar] [CrossRef] [PubMed]
- Benson, T.E.; Harris, M.S.; Choi, G.H.; Cialdella, J.I.; Herberg, J.T.; Martin, J.P., Jr.; Baldwin, E.T. A structural variation for murb: X-ray crystal structure of staphylococcus aureus udp-n-acetylenolpyruvylglucosamine reductase (murb). Biochemistry 2001, 40, 2340–2350. [Google Scholar] [CrossRef] [PubMed]
- Dolzhenko, A.V.; Pastorin, G.; Dolzhenko, A.V.; Chui, W.K. An aqueous medium synthesis and tautomerism study of 3(5)-amino-1,2,4-triazoles. Tetrahedron Lett. 2009, 50, 2124–2128. [Google Scholar] [CrossRef]
- Bera, H.; Tan, B.J.; Sun, L.; Dolzhenko, A.V.; Chui, W.-K.; Chiu, G.N.C. A structure–activity relationship study of 1,2,4-triazolo [1,5-a] [1,3,5]triazin-5,7-dione and its 5-thioxo analogues on anti-thymidine phosphorylase and associated anti-angiogenic activities. Eur. J. Med. Chem. 2013, 67, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, Y.; Chen, S.; Chen, B.; Jia, Y.; Zeng, Z. Synthesis and biological activities of schiff bases of 3-amino-1h-1, 2, 4-triazole. Chin. J. Chem. 2009, 27, 949–952. [Google Scholar] [CrossRef]
- Constantine, K.L.; Mueller, L.; Goldfarb, V.; Wittekind, M.; Metzler, W.J.; Yanchunas, J., Jr. Characterization of nadp+ binding to perdeuterated murb: Backbone atom nmr assignments and chemical-shift changes. J. Mol. Biol. 1997, 267, 1223–1246. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.V.; Patel, M.N. Insilico design, synthesis and evaluation of murb inhibitors as antibacterial agents. Int. J. Pharm. Technol. 2011, 3, 3064–3082. [Google Scholar]
- Arendrup, M.C.; Cuenca-Estrella, M.; Lass-Flörl, C.; Hope, W. Eucast technical note on the eucast definitive document edef 7.2: Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts edef 7.2 (eucast-afst). Clin. Microbiol. Infect. 2012, 18, E246–E247. [Google Scholar] [CrossRef] [PubMed]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Determination of minimum inhibitory concentrations (mics) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, ix–xv. [Google Scholar]
- Profeta, S., Jr.; Allinger, N. Molecular mechanics calculations on aliphatic amines. J. Am. Chem. Soc. 1985, 107, 1907–1918. [Google Scholar] [CrossRef]
- Allinger, N.L. Conformational analysis. 130. Mm2. A hydrocarbon force field utilizing v1 and v2 torsional terms. J. Am. Chem. Soc. 1977, 99, 8127–8134. [Google Scholar] [CrossRef]
- Labute, P.; Williams, C.; Feher, M.; Sourial, E.; Schmidt, J.M. Flexible alignment of small molecules. J. Med. Chem. 2001, 44, 1483–1490. [Google Scholar] [CrossRef] [PubMed]
- Kearsley, S.K.; Smith, G.M. An alternative method for the alignment of molecular structures: Maximizing electrostatic and steric overlap. Tetrahedron Comput. Methodol. 1990, 3, 615–633. [Google Scholar] [CrossRef]
- Sample Availability: Not available.











| Compound No. | cLogP a | Gram-negative Bacteria Gram-Positive Bacteria | Fungi | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli ATCC 25922 | Proteus vulgaris atcc 6380 | Pseudomonas aeruginosa ATCC 27857 | Staphylococcus aureus ATCC 29213 | Enterococcus faecalis ATCC 29212 | Bacillus subtilis ATCC 10400 | Mycobacterium fortuitum ATCC 6841 | Candida albicans ATCC 2091 | ||||||||||
| IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | ||
| 7c | 5.12 | 17 | 64 | Nil | ND | Nil | ND | 16 | 128 | Nil | ND | Nil | ND | 20 | 64 | Nil | ND |
| 7d | 4.55 | Nil | ND | Nil | ND | Nil | ND | Nil | ND | Nil | ND | Nil | ND | 9 | >128 | Nil | ND |
| 7e | 4.15 | Nil | ND | Nil | ND | Nil | ND | Nil | ND | Nil | ND | Nil | ND | 9 | >128 | Nil | ND |
| 7f | 4.32 | Nil | ND | Nil | ND | Nil | ND | Nil | ND | Nil | ND | Nil | ND | 10 | >128 | Nil | ND |
| 7g | 4.41 | Nil | ND | Nil | ND | Nil | ND | Nil | ND | Nil | ND | Nil | ND | 9 | >128 | Nil | ND |
| 7h | 3.84 | 30 | 16 | Nil | ND | 20 | 32 | 21 | 8 | 19 | 16 | 15 | 16 | 25 | 32 | 20 | 4 |
| 7i | 3.44 | 19 | 64 | Nil | ND | 18 | 32 | 14 | 128 | Nil | ND | Nil | ND | 12 | >128 | 19 | 16 |
| 7j | 3.62 | 22 | 32 | Nil | ND | 16 | 32 | 16 | 16 | Nil | ND | Nil | ND | 22 | 64 | 18 | 16 |
| 7k | 4.19 | 17 | 64 | Nil | ND | 17 | 32 | 15 | 128 | Nil | ND | Nil | ND | Nil | ND | 16 | 16 |
| 7l | 4.91 | 23 | 32 | Nil | ND | 18 | 32 | 17 | 64 | Nil | ND | Nil | ND | Nil | ND | 20 | 16 |
| CIPRO | 27 | ≤0.25 | 25 | ≤0.25 | 29 | ≤0.25 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| MOX | ND | ND | ND | ND | ND | ND | 28 | <0.002 | 25 | 0.5 | 27 | 0.094 | 26 | 0.125 | ND | ND | |
| FL | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 28 | 0.5 | |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, S.; Zayed, M.F.; El-Messery, S.M.; Al-Agamy, M.H.; Abdel-Rahman, H.M. Design, Synthesis, Antimicrobial Evaluation and Molecular Modeling Study of 1,2,4-Triazole-Based 4-Thiazolidinones. Molecules 2016, 21, 568. https://doi.org/10.3390/molecules21050568
Ahmed S, Zayed MF, El-Messery SM, Al-Agamy MH, Abdel-Rahman HM. Design, Synthesis, Antimicrobial Evaluation and Molecular Modeling Study of 1,2,4-Triazole-Based 4-Thiazolidinones. Molecules. 2016; 21(5):568. https://doi.org/10.3390/molecules21050568
Chicago/Turabian StyleAhmed, Sahar, Mohamed F. Zayed, Shahenda M. El-Messery, Mohamed H. Al-Agamy, and Hamdy M. Abdel-Rahman. 2016. "Design, Synthesis, Antimicrobial Evaluation and Molecular Modeling Study of 1,2,4-Triazole-Based 4-Thiazolidinones" Molecules 21, no. 5: 568. https://doi.org/10.3390/molecules21050568
APA StyleAhmed, S., Zayed, M. F., El-Messery, S. M., Al-Agamy, M. H., & Abdel-Rahman, H. M. (2016). Design, Synthesis, Antimicrobial Evaluation and Molecular Modeling Study of 1,2,4-Triazole-Based 4-Thiazolidinones. Molecules, 21(5), 568. https://doi.org/10.3390/molecules21050568