Rosmarinic Acid Methyl Ester Inhibits LPS-Induced NO Production via Suppression of MyD88- Dependent and -Independent Pathways and Induction of HO-1 in RAW 264.7 Cells
Abstract
:1. Introduction
2. Results
2.1. Effect of RAME on Cytotoxicity
2.2. Effect of RAME on NO Production and iNOS Protein Expression Levels
2.3. Effect of RAME on NF-κB Activation in LPS-Treated RAW 264.7 Cells
2.4. Effect of RAME on the Expression of Pro-Inflammatory Cytokines in LPS-Treated RAW 264.7 Cells
2.5. Effect of RAME on the IFN-β Pathway in LPS-Treated RAW 264.7 Cells
2.6. Effect of RAME on HO-1 Induction and Nrf2 Activation
3. Discussion
4. Materials and Methods
4.1. General Procedures
4.2. Materials
4.3. Extraction and Isolation
4.4. Cell Culture
4.5. Cytotoxicity Assay
4.6. Nitrite Assay
4.7. Western Blotting
4.8. Quantitative Real-Time Polymerase Chain Reaction
4.9. Luciferase Assay
4.10. Measurement of IFN-β and MCP-1 Levels
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kim, K.N.; Heo, S.J.; Yoon, W.J.; Kang, S.M.; Ahn, G.; Yi, T.H.; Jeon, Y.J. Fucoxanthin inhibits the inflammatory response by suppressing the activation of NF-κB and MAPKs in lipopolysaccharide-induced RAW 264.7 macrophages. Eur. J. Pharmacol. 2010, 649, 369–375. [Google Scholar] [PubMed]
- Guzik, T.J.; Korbut, R.; Adamek-Guzik, T. Nitric oxide and superoxide in inflammation and immune regulation. J. Physiol. Pharmacol. 2003, 54, 469–487. [Google Scholar] [PubMed]
- Nathan, C. Points of control in inflammation. Nature 2002, 420, 846–852. [Google Scholar] [PubMed]
- Walsh, N.C.; Crotti, T.N.; Goldring, S.R.; Gravallese, E.M. Rheumatic diseases: The effects of inflammation on bone. Immunol. Rev. 2005, 208, 228–251. [Google Scholar] [PubMed]
- Lee, H.J.; Hyun, E.A.; Yoon, W.J.; Kim, B.H.; Rhee, M.H.; Kang, H.K.; Cho, J.Y.; Yoo, E.S. In vitro anti-inflammatory and anti-oxidative effects of Cinnamomum camphora extracts. J. Ethnopharmacol. 2006, 103, 208–216. [Google Scholar] [PubMed]
- Lowenstein, C.J.; Hill, S.L.; Lafond-Walker, A.; Wu, J.; Allen, G.; Landavere, M.; Rose, N.R.; Herskowitz, A. Nitric oxide inhibits viral replication in murine myocarditis. J. Clin. Investig. 1996, 97, 1837–1843. [Google Scholar] [PubMed]
- Palmer, R.M.; Ashton, D.S.; Moncada, S. Vascular endothelial cells synthesize nitric oxide from l-arginine. Nature 1988, 333, 664–666. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.Y.; Dawson, V.L.; Dawson, T.M. Neurobiology of nitric oxide. Crit. Rev. Neurobiol. 1996, 10, 291–316. [Google Scholar] [CrossRef] [PubMed]
- Moncada, S.; Palmer, R.M.; Higgs, E.A. Nitric oxide: Physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 1991, 43, 109–142. [Google Scholar] [PubMed]
- Mordan, L.J.; Burnett, T.S.; Zhang, L.X.; Tom, J.; Cooney, R.V. Inhibitors of endogenous nitrogen oxide formation block the promotion of neoplastic transformation in C3H10T1/2 fibroblasts. Carcinogenesis 1993, 14, 1555–1559. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, H.; Bartsch, H. Chronic infections and inflammatory processes as cancer risk factors: Possible role of nitric oxide in carcinogenesis. Mutat. Res. 1994, 305, 253–264. [Google Scholar] [CrossRef]
- Kröncke, K.D.; Fehsel, K.; Kolb-Bachofen, V. Inducible nitric oxide synthase in human diseases. Clin. Exp. Immunol. 1998, 113, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.H.; Kim, E.S.; Jung, S.H.; Nho, C.W.; Lee, J.K. Tectorigenin inhibits IFN-gamma/LPS-induced inflammatory responses in murine macrophage RAW 264.7 cells. Arch. Pharm. Res. 2008, 31, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, E.M.; Hock, T.; Hill-Kapturczak, N.; Agarwal, A. The story so far: Molecular regulation of the heme oxygenase-1 gene in renal injury. Am. J. Physiol. Renal Physiol. 2004, 86, F425–F441. [Google Scholar] [CrossRef] [PubMed]
- Camhi, S.L.; Alam, J.; Wiegand, G.W.; Chin, B.Y.; Choi, A.M. Transcriptional activation of the HO-1 gene by lipopolysaccharide is mediated by 5′ distal enhancers: Role of reactive oxygen intermediates and AP-1. Am. J. Respir. Cell Mol. Biol. 1998, 18, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.H.; Lee, H.J.; Park, Y.D.; Choi, D.S.; Kim, D.S.; Kang, S.Y.; Seo, K.I.; Jeong, I.Y. Isoegomaketone inhibits lipopolysaccharide-induced nitric oxide production in RAW 264.7 macrophages through the heme oxygenase-1 induction and inhibition of the interferon-beta-STAT-1 pathway. J. Agric. Food Chem. 2010, 58, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Alam, J.; Stewart, D.; Touchard, C.; Boinapally, S.; Choi, A.M.; Cook, J.L. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 1999, 274, 26071–26078. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.A.; Johnson, D.A.; Kraft, A.D.; Calkins, M.J.; Jakel, R.J.; Vargas, M.R.; Chen, P.C. The Nrf2-ARE pathway: An indicator and modulator of oxidative stress in neurodegeneration. Ann. N. Y. Acad. Sci. 2008, 1147, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Steigel, A.; Reininger, E.; Bauer, R. Two new prenylated 3-benzoxepin derivatives ascyclooxygenase inhibitors from Perilla frutescens var. acuta. J. Nat. Prod. 2000, 63, 403–405. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Ohto, Y.; Murakami, A.; Ohigashi, H. Superoxide scavenging activity of rosmarinic acid from Perilla frutescens Britton Var. acuta f. Viridis. J. Agric. Food Chem. 1998, 46, 4545–4550. [Google Scholar] [CrossRef]
- Osakabe, N.; Yasuda, A.; Natsume, M.; Sanbongi, C.; Kato, Y.; Osawa, T.; Yoshikawa, T. Rosmarinic acid, a major polyphenolic component of Perilla frutescens, reduces lipopolysaccharide (lps)-induced liver injury in d-galactosamine (d-galn) sensitized mice. Free Radic Biol. Med. 2002, 33, 798–806. [Google Scholar] [CrossRef]
- Makino, T.; Furuta, Y.; Wakushima, H.; Fujii, H.; Saito, K.; Kano, Y. Anti-allergic effect of Perilla frutescens and its active constituents. Phytother. Res. 2003, 17, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Akyoshis, S.; Takashi, S.; Ryosuke, A.; Keisuks, S.; Kouta, N.; Chie, O.; Yoh, H.; Sadao, K.; Ueda, H.; Yamazaki, M. Toward Use of the Leaves of Perilla frutescens (L.) Britton var. Acuta Kudo (red perilla) with Japanese Dietary Pickled Plum (Umeboshi). J. Oleo Sci. 2006, 55, 413–422. [Google Scholar]
- Zhu, F.; Xu, Z.; Yonekura, L.; Yang, R.; Tamura, H. Antiallergic activity of rosmarinic acid esters is modulated by hydrophobicity, and bulkiness of alkyl side chain. Biosci. Biotechnol. Biochem. 2015, 79, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.; Vincent, R.; Séverine, M.; Murielle, B.; Annie, S.V.; Céline, R.; Sevser, S.; François, B.; Christel, N.; Thierry, H. Rosmarinic Acid and Its Methyl Ester as Antimicrobial Components of the Hydromethanolic Extract of Hyptis atrorubens Poit. (Lamiaceae). ECAM 2013, 2013. [Google Scholar] [CrossRef]
- Lim, H.J.; Woo, K.W.; Lee, K.R.; Lee, S.K.; Kim, H.P. Inhibition of Proinflammatory Cytokine Generation in Lung Inflammation by the Leaves of Perilla frutescens and Its Constituents. Biomol. Ther. (Seoul) 2014, 22, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.S.; Kim, W.J.; Kim, J.J.; Kim, T.J.; Ye, S.K.; Song, M.D.; Kang, H.; Kim, D.W.; Moon, W.K.; Lee, K.H. Gold nanoparticles attenuate LPS-induced NO production through the inhibition of NF-kappaB and IFN-beta/STAT1 pathways in RAW 264.7 cells. Nitric Oxide 2010, 23, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.T.; Ignarro, L.J. Lipopolysaccharide-induced expression of interferon-beta mediates the timing of inducible nitric-oxide synthase induction in RAW 264.7 macrophages. J. Biol. Chem. 2001, 276, 47950–47957. [Google Scholar] [PubMed]
- Halasi, M.; Wang, M.; Chavan, T.S.; Gaponenko, V.; Hay, N.; Gartel, A.L. ROS inhibitor N-acetyl-l-cysteine antagonizes the activity of proteasome inhibitors. Biochem. J. 2013, 454, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Tsuji, M.; Inazu, M.; Egashira, T.; Matsumiya, T. Rosmarinic acid and caffeic acid produce antidepressive-like effect in the forced swimming test in mice. Eur. J. Pharmacol. 2002, 449, 261–267. [Google Scholar] [CrossRef]
- Selvendiran, K.; Koga, H.; Ueno, T.; Yoshida, T.; Maeyama, M.; Torimura, T.; Yano, H.; Kojiro, M.; Sata, M. Luteolin promotes degradation in signal transducer and activator of transcription 3 in human hepatoma cells: An implication for the antitumor potential of flavonoids. Cancer Res. 2006, 66, 4826–4834. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Xu, Y.; Wen, X.; Nie, H.; Hu, T.; Yang, X.; Chu, X.; Yang, J.; Deng, X.; He, J. Rosmarinic Acid Attenuates Airway Inflammation and Hyperresponsiveness in a Murine Model of Asthma. Molecules 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Hauck, C.; Yum, M.Y.; Rizshsky, L.; Widrlechner, M.P.; McCoy, J.A.; Murphy, P.A.; Dixon, P.M.; Nikolau, B.J.; Birt, D.F. Rosmarinic acid in Prunella vulgaris ethanol extract inhibits lipopolysaccharide-induced prostaglandin E2 and nitric oxide in RAW 264.7 mouse macrophages. J. Agric. Food Chem. 2009, 57, 10579–10589. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.D.; Kang, M.A.; Lee, H.J.; Jin, C.H.; Choi, D.S.; Kim, D.S.; Kang, S.Y.; Byun, M.W.; Jeong, I.Y. Inhibition of an inducible nitric oxide synthase expression by a hexane extract from Perilla frutescens cv. chookyoupjaso mutant induced by mutagenesis with gamma-ray. J. Radiat. Ind. 2009, 3, 13–18. [Google Scholar]
- Kawai, T.; Akira, S. TLR signaling. Cell Death Differ. 2006, 13, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Sugiyama, M.; Yamamoto, M.; Watanabe, Y.; Kawai, T.; Takeda, K.; Akira, S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 2003, 171, 4304–4310. [Google Scholar] [PubMed]
- Covert, M.W.; Leung, T.H.; Gaston, J.E.; Baltimore, D. Achieving stability of lipopolysaccharide -induced NF-kappaB activation. Science 2005, 309, 1854–1857. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.S.; Noh, Y.S.; Lee, Y.S.; Cho, Y.W.; Baek, N.I.; Choi, M.S.; Jeong, T.S.; Kang, E.; Chung, H.G.; Lee, K.T. Arvelexin from Brassica rapa suppresses NF-κB-regulated pro-inflammatory gene expression by inhibiting activation of IκB kinase. Br. J. Pharmacol. 2011, 164, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Yoon, W.J.; Moon, J.Y.; Song, G.; Lee, Y.K.; Han, M.S.; Lee, J.S.; Ihm, B.S.; Lee, W.J.; Lee, N.H.; Hyun, C.G. Artemisia fukudo essential oil attenuates LPS-induced inflammation by suppressing NF-kappaB and MAPK activation in RAW 264.7 macrophages. Food Chem. Toxicol. 2010, 48, 1222–1229. [Google Scholar] [CrossRef] [PubMed]
- Reddy, D.B.; Reddanna, P. Chebulagic acid (CA) attenuates LPS-induced inflammation by suppressing NF-kappaB and MAPK activation in RAW 264.7 macrophages. Biochem. Biophys. Res. Commun. 2009, 381, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Han, J.M.; Lee, W.S.; Kim, J.R.; Son, J.; Kwon, O.H.; Lee, H.J.; Lee, J.J.; Jeong, T.S. Effect of 5-O-Methylhirsutanonol on nuclear factor-kappaB-dependent production of NO and expression of iNOS in lipopolysaccharide-induced RAW 264.7 cells. J. Agric. Food Chem. 2008, 56, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.L.; Zhu, Y.F.; Hu, M.M.; Wang, D.M.; Xu, X.J.; Lu, C.J.; Zhu, W. Optimization of astilbin extraction from the rhizome of Smilax glabra, and evaluation of its anti-inflammatory effect and probable underlying mechanism in lipopolysaccharide-induced RAW 264.7 macrophages. Molecules 2015, 20, 625–644. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Hirahashi, J.; Cullere, X.; Mayadas, T.N. Elucidation of molecular events leading to neutrophil apoptosis following phagocytosis: Cross-talk between caspase 8, reactive oxygen species, and MAPK/ERK activation. J. Biol. Chem. 2003, 278, 28443–28454. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Holst, O.; Di, L.F.; Callaghan, M.; Nurisso, A.; D’Errico, G.; Zamyatina, A.; Peri, F.; Berisio, R.; Jerala, R.; et al. Chemistry of lipid A: At the heart of innate immunity. Chemistry 2015, 21, 500–519. [Google Scholar] [CrossRef] [PubMed]
- Canes, L.; Borghi, C.; Ciacci, C.; Fabbri, R.; Vergani, L.; Gallo, G. Bisphenol-A alters gene expression and functional parameters in molluscan hepatopancreas. Mol. Cell Endocrinol. 2007, 276, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.









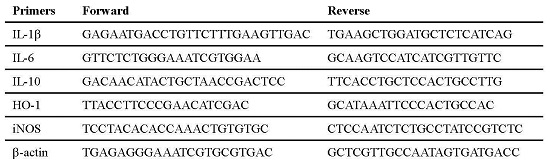
| Primers | Forward | Reverse |
|---|---|---|
| IL-1β | GAGAATGACCTGTTCTTTGAAGTTGAC | TGAAGCTGGATGCTCTCATCAG |
| IL-6 | GTTCTCTGGGAAATCGTGGAA | GCAAGTCCATCATCGTTGTTC |
| IL-10 | GACAACATACTGCTAACCGACTCC | TTCACCTGCTCCACTGCCTTG |
| HO-1 | TTACCTTCCCGAACATCGAC | GCATAAATTCCCACTGCCAC |
| iNOS | TCCTACACACCAAACTGTGTGC | CTCCAATCTCTGCCTATCCGTCTC |
| β-actin | TGAGAGGGAAATCGTGCGTGAC | GCTCGTTGCCAATAGTGATGACC |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
So, Y.; Lee, S.Y.; Han, A.-R.; Kim, J.-B.; Jeong, H.G.; Jin, C.H. Rosmarinic Acid Methyl Ester Inhibits LPS-Induced NO Production via Suppression of MyD88- Dependent and -Independent Pathways and Induction of HO-1 in RAW 264.7 Cells. Molecules 2016, 21, 1083. https://doi.org/10.3390/molecules21081083
So Y, Lee SY, Han A-R, Kim J-B, Jeong HG, Jin CH. Rosmarinic Acid Methyl Ester Inhibits LPS-Induced NO Production via Suppression of MyD88- Dependent and -Independent Pathways and Induction of HO-1 in RAW 264.7 Cells. Molecules. 2016; 21(8):1083. https://doi.org/10.3390/molecules21081083
Chicago/Turabian StyleSo, Yangkang, Seung Young Lee, Ah-Reum Han, Jin-Baek Kim, Hye Gwang Jeong, and Chang Hyun Jin. 2016. "Rosmarinic Acid Methyl Ester Inhibits LPS-Induced NO Production via Suppression of MyD88- Dependent and -Independent Pathways and Induction of HO-1 in RAW 264.7 Cells" Molecules 21, no. 8: 1083. https://doi.org/10.3390/molecules21081083
APA StyleSo, Y., Lee, S. Y., Han, A. -R., Kim, J. -B., Jeong, H. G., & Jin, C. H. (2016). Rosmarinic Acid Methyl Ester Inhibits LPS-Induced NO Production via Suppression of MyD88- Dependent and -Independent Pathways and Induction of HO-1 in RAW 264.7 Cells. Molecules, 21(8), 1083. https://doi.org/10.3390/molecules21081083